Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
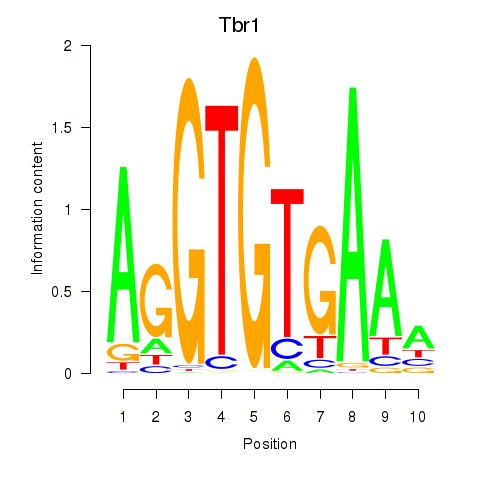
Results for Tbr1
Z-value: 1.92
Transcription factors associated with Tbr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbr1
|
ENSMUSG00000035033.16 | Tbr1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbr1 | mm39_v1_chr2_+_61634797_61634890 | -0.57 | 2.0e-07 | Click! |
Activity profile of Tbr1 motif
Sorted Z-values of Tbr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 2.7 | 8.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 2.6 | 12.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.2 | 6.7 | GO:0002215 | defense response to nematode(GO:0002215) |
| 2.2 | 6.5 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 2.0 | 6.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.9 | 5.6 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 1.8 | 7.2 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.6 | 4.8 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.6 | 6.2 | GO:2001189 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 1.5 | 4.5 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 1.5 | 4.5 | GO:0009233 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.5 | 4.4 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 1.4 | 8.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.3 | 4.0 | GO:2000566 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 1.3 | 5.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.3 | 7.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.3 | 3.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.3 | 10.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.2 | 6.2 | GO:2001274 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 1.2 | 3.7 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.2 | 4.9 | GO:0000239 | pachytene(GO:0000239) |
| 1.2 | 6.0 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.2 | 4.8 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 1.2 | 3.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 1.2 | 4.6 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 1.2 | 3.5 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.1 | 5.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.1 | 3.3 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 1.1 | 3.3 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 1.1 | 18.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 1.0 | 4.1 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 1.0 | 3.0 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.0 | 5.9 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.0 | 1.9 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.9 | 2.8 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.9 | 8.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.9 | 2.7 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.9 | 4.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.9 | 4.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.9 | 2.6 | GO:0002588 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.9 | 2.6 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.9 | 0.9 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.8 | 1.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.8 | 2.5 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.8 | 3.3 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.8 | 1.6 | GO:0002856 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.8 | 6.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.8 | 2.3 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.8 | 15.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.7 | 2.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.7 | 5.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.7 | 12.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.7 | 2.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.7 | 5.7 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.7 | 5.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.7 | 2.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.7 | 2.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.7 | 4.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.7 | 3.4 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.7 | 2.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.7 | 3.3 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.6 | 1.9 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.6 | 1.9 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.6 | 6.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.6 | 3.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.6 | 3.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.6 | 3.7 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.6 | 1.8 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.6 | 8.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.6 | 3.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.6 | 2.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 3.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.6 | 6.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.6 | 9.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 1.7 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.6 | 6.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.6 | 6.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.6 | 3.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.5 | 1.6 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.5 | 3.8 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.5 | 11.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.5 | 12.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.5 | 3.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 10.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.5 | 2.7 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.5 | 2.6 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.5 | 2.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.5 | 1.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.5 | 7.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.5 | 3.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 8.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.5 | 1.5 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.5 | 2.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.5 | 2.5 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.5 | 3.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.5 | 6.2 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.5 | 3.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.5 | 7.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.5 | 4.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.4 | 1.8 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 4.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.4 | 15.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.4 | 2.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.4 | 1.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.4 | 6.5 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.4 | 4.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.4 | 6.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 1.6 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.4 | 3.9 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.4 | 1.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.4 | 1.1 | GO:0046379 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.4 | 6.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.4 | 4.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.4 | 6.7 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.4 | 2.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.4 | 1.8 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 1.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.4 | 2.9 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.4 | 3.5 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.3 | 2.1 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.3 | 5.9 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.3 | 3.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.3 | 5.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 24.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.3 | 1.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 4.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 3.3 | GO:0051324 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.3 | 5.8 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.3 | 1.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 46.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 1.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.3 | 8.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 7.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 3.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 15.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 1.8 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.3 | 1.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 4.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 2.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 2.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 5.7 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.3 | 2.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 2.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.3 | 2.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 2.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 1.4 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.3 | 0.8 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.3 | 1.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.3 | 3.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 10.7 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.3 | 0.8 | GO:0036088 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.3 | 0.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.3 | 1.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 | 3.4 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.3 | 1.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 1.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 0.7 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 2.6 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 2.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 4.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.4 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.2 | 3.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 1.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 0.9 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 3.9 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 1.5 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.2 | 3.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.2 | 0.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 1.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 4.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 1.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 5.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 3.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 8.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 8.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 2.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 4.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.2 | 4.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 2.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 3.6 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 4.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 1.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 5.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 20.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 3.7 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.2 | 2.2 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 1.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 2.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.2 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 62.8 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.2 | 37.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 1.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 3.3 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.2 | 4.3 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.2 | 2.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 2.0 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.0 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 2.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 4.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.2 | 1.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 4.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 6.5 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.1 | 5.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 1.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.4 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 2.9 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 1.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.8 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 2.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 1.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.9 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 6.2 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.1 | 1.7 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 0.9 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.9 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 5.7 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 3.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 3.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 9.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.4 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 2.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 2.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 1.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 4.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 7.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 3.3 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.1 | 1.7 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 3.4 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 3.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 3.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.4 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 3.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 10.6 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.5 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.9 | GO:2001199 | regulation of dendritic cell differentiation(GO:2001198) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 2.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 1.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 1.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 2.3 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 3.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 1.8 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.7 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 17.6 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.1 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 1.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 1.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 2.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 2.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.6 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 4.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 3.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 1.5 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 2.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.2 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.3 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.8 | GO:0070918 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.9 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 1.7 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.7 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 2.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.6 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 2.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.5 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 2.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.6 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.2 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 1.6 | GO:0055123 | digestive system development(GO:0055123) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.6 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.5 | 17.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 2.1 | 12.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.0 | 30.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.5 | 4.5 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 1.5 | 4.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 1.2 | 4.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.1 | 3.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 1.1 | 3.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 1.1 | 6.4 | GO:0071547 | piP-body(GO:0071547) |
| 1.0 | 3.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 1.0 | 11.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.9 | 6.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 10.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 3.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 13.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 9.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.7 | 3.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.7 | 2.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.7 | 13.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.6 | 1.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.6 | 10.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.6 | 1.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.5 | 8.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.5 | 77.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.5 | 4.5 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 2.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.5 | 9.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 5.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.5 | 15.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 3.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 6.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 6.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 3.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 3.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 13.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 5.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 12.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 1.9 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.4 | 3.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 46.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 10.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.4 | 3.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 3.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 10.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 3.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 2.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 3.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 1.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 3.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 5.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 1.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 1.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 1.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 12.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 4.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 1.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 3.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 1.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 6.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 0.6 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 2.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 3.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 29.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 3.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 3.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 11.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 5.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 3.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 3.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 2.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 3.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 2.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 2.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 2.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 39.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 2.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 5.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 2.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 6.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 29.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 2.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 3.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 6.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 4.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 5.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 3.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 3.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 6.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.0 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 2.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 6.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 2.1 | 6.2 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.9 | 11.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.7 | 8.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 1.5 | 4.5 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.4 | 7.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.4 | 4.3 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 1.2 | 6.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.2 | 3.7 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 1.2 | 6.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.2 | 5.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.1 | 3.4 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 1.1 | 3.4 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 1.1 | 6.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.0 | 4.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 1.0 | 4.0 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.9 | 2.6 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.9 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.9 | 4.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.8 | 3.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.8 | 3.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 3.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.8 | 7.3 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.8 | 10.4 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.8 | 6.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.7 | 4.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.7 | 2.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.7 | 2.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 4.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.7 | 3.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.7 | 8.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 2.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.6 | 6.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.6 | 2.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.6 | 3.7 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.6 | 13.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 3.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.5 | 2.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.5 | 2.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 9.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.5 | 1.6 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.5 | 2.0 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 0.5 | 1.5 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.5 | 1.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.5 | 1.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 3.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.4 | 4.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 1.8 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 1.8 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 24.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.4 | 1.2 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.4 | 2.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 2.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.4 | 3.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 2.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 1.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 46.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 6.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.4 | 6.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 2.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 1.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 1.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.3 | 2.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 5.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 1.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 2.9 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 8.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 2.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 3.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 2.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 3.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 2.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 5.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 4.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 1.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 2.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 4.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 1.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 1.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.3 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 2.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 7.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 4.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 6.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.7 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.2 | 5.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 2.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 6.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 3.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 1.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.2 | 4.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 1.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 4.6 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.2 | 3.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.8 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 1.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 10.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 5.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 2.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.2 | 1.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 0.9 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.2 | 1.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 3.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 9.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 2.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 5.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 3.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 3.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.7 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 2.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 4.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 3.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 5.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.7 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 7.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 3.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 1.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 3.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 5.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 1.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 2.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.1 | 2.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 3.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.5 | GO:0023023 | MHC protein complex binding(GO:0023023) |
| 0.1 | 1.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 7.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 2.8 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 3.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.8 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 19.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 4.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 4.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 2.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 3.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 4.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 7.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 2.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 4.4 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 3.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 15.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 3.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 27.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 3.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 3.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.6 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.9 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 8.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.5 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 35.0 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.1 | GO:0035478 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) chylomicron binding(GO:0035478) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 19.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 10.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 16.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 3.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 13.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 3.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 2.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 3.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 8.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 7.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 5.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 17.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 6.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 4.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 11.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 0.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 2.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 4.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.8 | 16.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.7 | 8.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 1.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.4 | 2.7 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.4 | 21.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 5.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 7.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 7.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 3.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 3.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 2.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 2.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 2.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 2.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 2.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 4.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 1.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 5.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 2.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 5.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 4.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 14.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 3.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 5.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 6.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 5.5 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 1.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 14.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 5.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |