Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
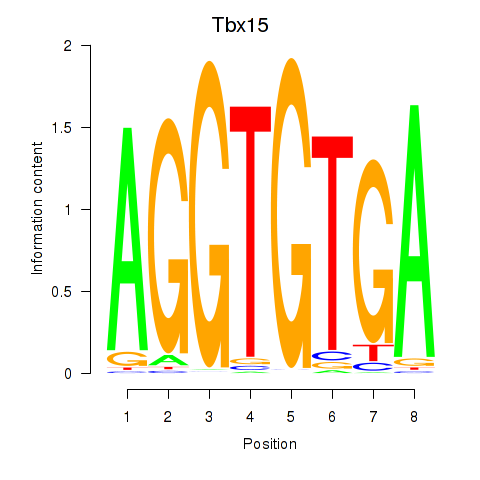
Results for Tbx15
Z-value: 1.27
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSMUSG00000027868.12 | Tbx15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx15 | mm39_v1_chr3_+_99147677_99147702 | -0.21 | 7.9e-02 | Click! |
Activity profile of Tbx15 motif
Sorted Z-values of Tbx15 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx15
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_95727267 | 13.63 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr6_+_115111860 | 8.41 |
ENSMUST00000169345.4
|
Syn2
|
synapsin II |
| chr2_-_13016570 | 8.36 |
ENSMUST00000061545.7
|
C1ql3
|
C1q-like 3 |
| chr4_-_129015682 | 7.76 |
ENSMUST00000139450.8
ENSMUST00000125931.9 ENSMUST00000116444.10 |
Hpca
|
hippocalcin |
| chr3_+_75464837 | 7.44 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr6_+_7555053 | 7.14 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr7_-_4607040 | 6.75 |
ENSMUST00000166650.3
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr6_+_115111872 | 6.65 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chrX_+_7786061 | 6.63 |
ENSMUST00000041096.4
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chrX_+_135567124 | 6.43 |
ENSMUST00000060904.11
ENSMUST00000113100.2 ENSMUST00000128040.2 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr3_+_96088467 | 6.22 |
ENSMUST00000035371.9
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr3_+_55369288 | 5.80 |
ENSMUST00000198412.5
ENSMUST00000199169.5 ENSMUST00000199702.5 ENSMUST00000198437.5 |
Dclk1
|
doublecortin-like kinase 1 |
| chr7_-_100307601 | 5.56 |
ENSMUST00000138830.2
ENSMUST00000107044.10 ENSMUST00000116287.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_-_116693849 | 5.53 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr7_+_27770655 | 5.49 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr15_+_103411689 | 5.38 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr1_+_153541412 | 5.13 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr6_+_78347844 | 5.07 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr2_+_25132941 | 5.06 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr6_+_78347636 | 4.88 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr11_+_69909245 | 4.85 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr7_-_74959010 | 4.82 |
ENSMUST00000165175.8
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr14_-_70415117 | 4.80 |
ENSMUST00000022681.11
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr18_+_82493284 | 4.79 |
ENSMUST00000047865.14
|
Mbp
|
myelin basic protein |
| chr11_+_69231589 | 4.71 |
ENSMUST00000218008.2
ENSMUST00000151617.3 |
Rnf227
|
ring finger protein 227 |
| chr7_-_45016138 | 4.67 |
ENSMUST00000211067.2
ENSMUST00000003961.16 |
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr7_-_100307571 | 4.62 |
ENSMUST00000107043.8
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr15_-_96540760 | 4.62 |
ENSMUST00000088452.11
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr2_-_180844582 | 4.61 |
ENSMUST00000016511.6
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr5_-_147244074 | 4.51 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr7_+_4122523 | 4.50 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr2_+_164810139 | 4.44 |
ENSMUST00000202479.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr8_-_4309257 | 4.44 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr15_-_98705791 | 4.38 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr11_+_66847446 | 4.38 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr6_-_24956296 | 4.34 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr3_+_98129463 | 4.30 |
ENSMUST00000029469.5
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr4_+_138559156 | 4.28 |
ENSMUST00000077582.14
|
Pla2g2a
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr6_-_127746390 | 4.18 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr1_+_153541020 | 4.18 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr19_-_46315543 | 4.17 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr4_-_119515978 | 4.15 |
ENSMUST00000106309.9
ENSMUST00000044426.8 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr6_+_114108190 | 4.00 |
ENSMUST00000032451.9
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
| chr1_-_172156884 | 3.89 |
ENSMUST00000062387.8
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr1_+_66507523 | 3.85 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr7_+_4122555 | 3.83 |
ENSMUST00000079415.12
|
Ttyh1
|
tweety family member 1 |
| chr3_+_55369384 | 3.80 |
ENSMUST00000200352.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr17_-_43813664 | 3.73 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chrX_+_40490005 | 3.61 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr16_-_9812787 | 3.60 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr3_+_55369149 | 3.60 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr6_-_68681962 | 3.59 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr19_+_28941292 | 3.57 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr8_-_41087793 | 3.55 |
ENSMUST00000173957.2
ENSMUST00000048898.17 ENSMUST00000174205.8 |
Mtmr7
|
myotubularin related protein 7 |
| chr7_+_25005510 | 3.52 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chrX_+_40490353 | 3.51 |
ENSMUST00000165288.2
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr7_+_91321500 | 3.50 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr11_+_69909659 | 3.46 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr9_-_4795519 | 3.45 |
ENSMUST00000212533.2
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr18_+_82493237 | 3.42 |
ENSMUST00000091789.11
ENSMUST00000114676.8 |
Mbp
|
myelin basic protein |
| chr1_+_83094481 | 3.41 |
ENSMUST00000027351.13
ENSMUST00000113437.9 ENSMUST00000186832.2 |
Ccl20
|
chemokine (C-C motif) ligand 20 |
| chr1_+_165957784 | 3.40 |
ENSMUST00000060833.14
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr14_-_76794103 | 3.40 |
ENSMUST00000064517.9
ENSMUST00000228055.2 |
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr9_-_15834052 | 3.40 |
ENSMUST00000217187.2
|
Fat3
|
FAT atypical cadherin 3 |
| chrX_-_149440388 | 3.39 |
ENSMUST00000151403.9
ENSMUST00000087253.11 ENSMUST00000112709.8 ENSMUST00000163969.8 ENSMUST00000087258.10 |
Tro
|
trophinin |
| chr7_-_97811525 | 3.39 |
ENSMUST00000107112.2
|
Capn5
|
calpain 5 |
| chr1_+_93789068 | 3.37 |
ENSMUST00000094663.4
|
Gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr1_-_172156828 | 3.37 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr6_-_131290790 | 3.31 |
ENSMUST00000049150.8
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr2_-_57004933 | 3.26 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr6_-_28831746 | 3.26 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr16_+_17093941 | 3.25 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr7_-_4606104 | 3.23 |
ENSMUST00000049113.14
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr6_-_69792108 | 3.20 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr9_+_27210500 | 3.16 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr12_+_88920169 | 3.16 |
ENSMUST00000057634.14
|
Nrxn3
|
neurexin III |
| chr12_-_113790741 | 3.16 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr9_+_59614877 | 3.15 |
ENSMUST00000128944.8
ENSMUST00000098661.10 |
Gramd2
|
GRAM domain containing 2 |
| chr5_+_120787253 | 3.15 |
ENSMUST00000156722.2
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr7_+_91321694 | 3.13 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr10_+_123099945 | 3.12 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr4_+_62398262 | 3.12 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr15_+_98532624 | 3.07 |
ENSMUST00000003442.9
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr15_-_75439013 | 3.07 |
ENSMUST00000156032.2
ENSMUST00000127095.8 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr10_+_103203552 | 3.07 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chr3_-_84128160 | 3.06 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr4_-_129132963 | 3.04 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr5_-_148336711 | 3.02 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_-_10282218 | 2.98 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr11_-_83959175 | 2.92 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr7_-_112968533 | 2.92 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chrX_+_133195974 | 2.92 |
ENSMUST00000037687.8
|
Tmem35a
|
transmembrane protein 35A |
| chr11_-_103247150 | 2.89 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr10_+_75402090 | 2.87 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_+_124124493 | 2.86 |
ENSMUST00000212880.2
|
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr1_-_57010921 | 2.86 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_-_130172998 | 2.84 |
ENSMUST00000120126.9
|
Serinc2
|
serine incorporator 2 |
| chr15_+_103411461 | 2.83 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr13_+_3974686 | 2.83 |
ENSMUST00000021639.8
|
Tubal3
|
tubulin, alpha-like 3 |
| chr2_+_28423367 | 2.81 |
ENSMUST00000113893.8
ENSMUST00000100241.10 |
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr7_-_78227518 | 2.79 |
ENSMUST00000195262.6
ENSMUST00000193002.6 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr4_+_115595610 | 2.78 |
ENSMUST00000106524.4
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr16_-_44153498 | 2.78 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr18_-_47466378 | 2.77 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_+_128736827 | 2.76 |
ENSMUST00000219317.2
|
Cd63
|
CD63 antigen |
| chr10_+_81228877 | 2.76 |
ENSMUST00000105322.9
|
Smim24
|
small integral membrane protein 24 |
| chr6_-_69741999 | 2.74 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr16_-_44153288 | 2.73 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr5_-_148336574 | 2.73 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_-_95602952 | 2.73 |
ENSMUST00000046461.9
|
Dok4
|
docking protein 4 |
| chr5_-_84565218 | 2.72 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr4_+_133788065 | 2.70 |
ENSMUST00000227683.2
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr2_+_21372338 | 2.70 |
ENSMUST00000055946.8
|
Gpr158
|
G protein-coupled receptor 158 |
| chr12_+_69954506 | 2.69 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr4_+_48045143 | 2.66 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr14_+_52254341 | 2.64 |
ENSMUST00000228408.2
ENSMUST00000227295.2 |
Tmem253
|
transmembrane protein 253 |
| chr8_+_14145848 | 2.63 |
ENSMUST00000152652.8
ENSMUST00000133298.8 |
Dlgap2
|
DLG associated protein 2 |
| chr11_+_97732108 | 2.61 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr4_-_150736554 | 2.61 |
ENSMUST00000117997.2
ENSMUST00000037827.10 |
Slc45a1
|
solute carrier family 45, member 1 |
| chr6_-_69800923 | 2.59 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr1_+_153541339 | 2.57 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chrX_-_58613428 | 2.55 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr1_+_34717230 | 2.55 |
ENSMUST00000159747.9
|
Arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_91013362 | 2.55 |
ENSMUST00000066420.12
|
Madd
|
MAP-kinase activating death domain |
| chr6_-_69835868 | 2.51 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr3_+_94385602 | 2.50 |
ENSMUST00000199884.5
ENSMUST00000198316.5 ENSMUST00000197558.5 |
Celf3
|
CUGBP, Elav-like family member 3 |
| chr1_+_72323526 | 2.46 |
ENSMUST00000027380.12
ENSMUST00000141783.2 |
Tmem169
|
transmembrane protein 169 |
| chr14_+_52254308 | 2.45 |
ENSMUST00000100638.4
|
Tmem253
|
transmembrane protein 253 |
| chr11_-_113540867 | 2.41 |
ENSMUST00000136392.8
ENSMUST00000125890.8 ENSMUST00000146031.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr11_+_98632696 | 2.36 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr6_+_68414401 | 2.36 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr1_+_165957909 | 2.35 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr16_+_21644692 | 2.33 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr3_-_129126362 | 2.32 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr12_+_88689638 | 2.30 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr7_-_126224848 | 2.28 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr3_-_81981946 | 2.28 |
ENSMUST00000029635.14
ENSMUST00000193597.2 |
Gucy1b1
|
guanylate cyclase 1, soluble, beta 1 |
| chr10_-_81308693 | 2.27 |
ENSMUST00000147524.3
ENSMUST00000119060.8 |
Celf5
|
CUGBP, Elav-like family member 5 |
| chr11_+_98632953 | 2.26 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr16_-_9812410 | 2.26 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr4_+_125384481 | 2.23 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr9_-_49710190 | 2.23 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chrX_-_156826262 | 2.23 |
ENSMUST00000026750.15
ENSMUST00000112513.2 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr4_+_42612121 | 2.21 |
ENSMUST00000178168.3
|
Gm10591
|
predicted gene 10591 |
| chr2_-_91013193 | 2.21 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chrX_+_99773523 | 2.18 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr7_-_105289515 | 2.18 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chrX_+_99773784 | 2.18 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr6_+_70549568 | 2.17 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr11_+_61967821 | 2.15 |
ENSMUST00000092415.9
ENSMUST00000201015.4 ENSMUST00000202744.4 ENSMUST00000201723.4 ENSMUST00000202179.2 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr11_+_53410552 | 2.15 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr5_+_146392371 | 2.14 |
ENSMUST00000238592.2
|
Wasf3
|
WASP family, member 3 |
| chr9_-_49710058 | 2.14 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr11_+_108814007 | 2.11 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr3_+_126391046 | 2.11 |
ENSMUST00000106401.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr16_+_93480061 | 2.10 |
ENSMUST00000039620.7
|
Cbr3
|
carbonyl reductase 3 |
| chr15_-_82796308 | 2.10 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr2_-_131987008 | 2.10 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr7_-_115837034 | 2.10 |
ENSMUST00000182834.8
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr15_-_79048674 | 2.09 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr6_+_61157279 | 2.09 |
ENSMUST00000126214.8
|
Ccser1
|
coiled-coil serine rich 1 |
| chr2_+_138120401 | 2.08 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr11_+_98632631 | 2.07 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr11_-_51748450 | 2.06 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr9_-_106762818 | 2.05 |
ENSMUST00000185707.2
|
Rbm15b
|
RNA binding motif protein 15B |
| chr3_-_158267771 | 2.03 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr12_-_113561594 | 2.03 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chrX_+_41591355 | 2.03 |
ENSMUST00000189753.7
|
Sh2d1a
|
SH2 domain containing 1A |
| chr4_+_53440389 | 2.02 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chrX_-_149440362 | 1.99 |
ENSMUST00000148604.2
|
Tro
|
trophinin |
| chr7_+_48438751 | 1.98 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr4_+_41903610 | 1.97 |
ENSMUST00000098128.4
|
Ccl21d
|
chemokine (C-C motif) ligand 21D |
| chr6_-_69753317 | 1.97 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr7_-_78228116 | 1.95 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr11_-_54853729 | 1.93 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr4_+_42255693 | 1.93 |
ENSMUST00000178864.3
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr6_-_94677118 | 1.92 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr19_+_23881821 | 1.92 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr1_-_87322443 | 1.91 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr14_-_119162736 | 1.91 |
ENSMUST00000004055.10
|
Dzip1
|
DAZ interacting protein 1 |
| chr6_+_70640233 | 1.90 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr8_+_79754980 | 1.89 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr19_-_42190589 | 1.88 |
ENSMUST00000018966.8
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr1_-_131204422 | 1.87 |
ENSMUST00000159195.2
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr6_+_108190163 | 1.87 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr5_+_64250268 | 1.85 |
ENSMUST00000087324.7
|
Pgm2
|
phosphoglucomutase 2 |
| chrX_+_138464065 | 1.85 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr3_+_126390951 | 1.85 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr2_+_68691902 | 1.85 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr7_+_126528016 | 1.85 |
ENSMUST00000032924.6
|
Kctd13
|
potassium channel tetramerisation domain containing 13 |
| chrX_+_139243012 | 1.85 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr7_+_46045862 | 1.83 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr7_-_24016020 | 1.82 |
ENSMUST00000108436.8
ENSMUST00000032673.15 |
Zfp94
|
zinc finger protein 94 |
| chr17_+_56277451 | 1.82 |
ENSMUST00000044216.8
ENSMUST00000225145.2 |
Shd
|
src homology 2 domain-containing transforming protein D |
| chr3_-_96812610 | 1.81 |
ENSMUST00000029738.14
|
Gpr89
|
G protein-coupled receptor 89 |
| chr18_-_39652468 | 1.78 |
ENSMUST00000237944.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr8_+_79755194 | 1.78 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr19_+_53128901 | 1.76 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr3_+_94385661 | 1.74 |
ENSMUST00000200342.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr2_-_172782089 | 1.74 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.6 | 7.8 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.1 | 8.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.8 | 7.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.7 | 6.7 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.6 | 4.8 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 1.5 | 4.4 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.2 | 6.9 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 1.1 | 3.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.1 | 4.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 1.1 | 3.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.0 | 12.6 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.9 | 4.7 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.8 | 4.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.8 | 11.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.7 | 3.0 | GO:0099541 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.7 | 5.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.7 | 2.1 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.7 | 2.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 | 1.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.6 | 1.9 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.6 | 8.3 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.6 | 8.2 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.6 | 1.7 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.6 | 4.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.5 | 2.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.5 | 1.5 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.5 | 5.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.5 | 1.5 | GO:0061193 | taste bud development(GO:0061193) |
| 0.5 | 2.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.5 | 0.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.4 | 1.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.4 | 2.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 6.2 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.4 | 2.8 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 6.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.4 | 3.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.4 | 1.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.4 | 3.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.4 | 3.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.4 | 2.6 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 9.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 2.9 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 1.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 2.3 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.3 | 6.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 2.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 1.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 1.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 6.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 2.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 2.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 2.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 1.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.3 | 1.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.3 | 0.8 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.3 | 3.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 3.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.3 | 4.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 1.8 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 1.7 | GO:0043545 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 2.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.2 | 1.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 4.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.3 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.2 | 1.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 6.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 2.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 0.8 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 2.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 10.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 2.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 0.8 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.2 | 3.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.2 | 0.7 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.2 | 0.6 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.2 | 0.9 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.2 | 0.3 | GO:0072223 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.2 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 0.8 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 3.1 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 3.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.2 | 1.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 1.1 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.2 | 0.5 | GO:0032262 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.2 | 4.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 2.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 2.3 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 3.0 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.1 | 7.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.6 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 4.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 0.7 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.1 | 2.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 2.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.4 | GO:0048822 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 2.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 4.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 2.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 11.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.5 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 1.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 6.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.5 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.3 | GO:0070100 | regulation of negative chemotaxis(GO:0050923) negative regulation of negative chemotaxis(GO:0050925) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 1.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.3 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.1 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 2.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.8 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 1.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.0 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 20.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 1.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 2.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 3.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 4.4 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 1.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 3.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 4.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.6 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:1990768 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.5 | GO:0031394 | JUN phosphorylation(GO:0007258) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 1.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 5.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 0.7 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 0.4 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 5.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 2.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 2.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.9 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 0.9 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 2.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 7.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.1 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.8 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 1.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 3.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 1.6 | GO:0015844 | monoamine transport(GO:0015844) |
| 0.0 | 0.4 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 1.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.8 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 0.4 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 3.2 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 10.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.6 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.4 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 1.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.5 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.3 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 2.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 2.6 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 2.1 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 2.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 3.9 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.4 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 2.4 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.7 | 8.5 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.5 | 13.6 | GO:1990357 | terminal web(GO:1990357) |
| 1.0 | 8.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.9 | 2.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.8 | 3.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.6 | 1.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.6 | 7.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 7.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 14.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.5 | 3.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.4 | 0.4 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.4 | 2.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 1.4 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 2.6 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.3 | 6.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 13.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 10.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 1.3 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.3 | 22.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 2.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 8.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 2.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 1.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 4.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 2.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 2.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 3.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 3.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 12.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 5.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.7 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 4.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 9.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 8.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 25.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 10.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 7.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 1.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 6.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 5.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 2.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 5.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 4.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 3.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 3.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 12.8 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.6 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 1.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 20.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.7 | 6.7 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.3 | 10.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.2 | 8.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.9 | 5.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.8 | 7.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.8 | 2.4 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.8 | 10.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.7 | 8.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.7 | 7.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.7 | 6.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.7 | 2.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.7 | 4.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.7 | 4.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.7 | 4.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.6 | 3.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 2.9 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.5 | 8.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 2.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 1.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 3.4 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.5 | 6.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 3.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 4.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.4 | 5.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 4.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 3.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 4.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 2.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 2.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 2.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 1.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 1.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 3.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 3.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 1.8 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 1.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 1.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 2.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 0.8 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.3 | 1.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.3 | 5.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 4.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 3.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 4.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.7 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 2.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 1.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 11.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 4.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.2 | 0.7 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.8 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 1.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.2 | 0.8 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 4.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.6 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 5.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 5.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 3.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.3 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 11.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 9.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 5.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 4.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 4.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 3.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 1.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 7.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 3.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 12.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 9.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 11.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 7.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 7.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 6.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 16.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 6.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 5.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 6.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 7.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 5.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 10.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 3.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 5.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.7 | 23.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 8.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 4.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 11.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 4.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 4.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 4.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 3.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 7.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 6.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 6.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 8.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 7.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 5.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 7.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.5 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |