Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
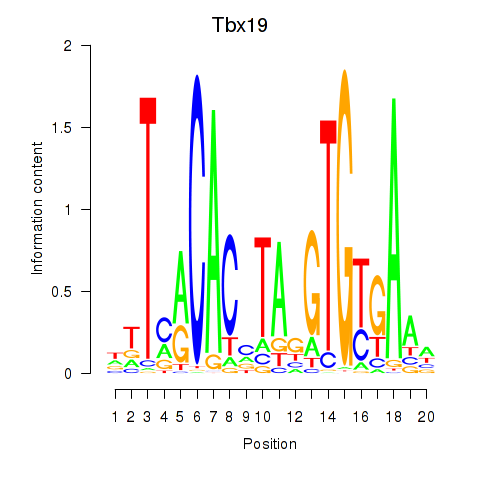
Results for Tbx19
Z-value: 1.37
Transcription factors associated with Tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx19
|
ENSMUSG00000026572.12 | Tbx19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx19 | mm39_v1_chr1_-_164988342_164988350 | -0.23 | 4.8e-02 | Click! |
Activity profile of Tbx19 motif
Sorted Z-values of Tbx19 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx19
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_24803336 | 19.61 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr19_-_40175709 | 17.27 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr3_+_138121245 | 14.99 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr13_+_4484305 | 14.85 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr10_-_93375832 | 13.15 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr4_-_60455331 | 13.00 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr8_-_110305672 | 12.49 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr17_-_18104182 | 11.81 |
ENSMUST00000061516.8
|
Fpr1
|
formyl peptide receptor 1 |
| chr9_-_103107495 | 11.65 |
ENSMUST00000035158.16
|
Trf
|
transferrin |
| chr7_-_19415301 | 10.46 |
ENSMUST00000150569.9
ENSMUST00000127648.4 ENSMUST00000003071.10 |
Gm44805
Apoc4
|
predicted gene 44805 apolipoprotein C-IV |
| chr3_+_94600863 | 9.69 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr11_-_59937302 | 9.39 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr9_-_103107460 | 9.21 |
ENSMUST00000165296.8
ENSMUST00000112645.8 |
Trf
|
transferrin |
| chr9_-_118986123 | 9.16 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr12_-_81014849 | 8.88 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr7_-_140535828 | 8.53 |
ENSMUST00000211129.2
|
Gm45717
|
predicted gene 45717 |
| chr7_-_140535899 | 8.45 |
ENSMUST00000081649.10
|
Ifitm2
|
interferon induced transmembrane protein 2 |
| chr12_-_81014755 | 8.02 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr19_+_56276375 | 7.25 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr19_+_56276343 | 7.17 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr8_-_41586713 | 6.86 |
ENSMUST00000155055.2
ENSMUST00000059115.13 ENSMUST00000145860.2 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr14_-_34032311 | 6.81 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr17_+_79934096 | 6.72 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr7_-_3723381 | 6.44 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr6_+_90442269 | 6.31 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr15_-_98626002 | 6.23 |
ENSMUST00000003445.8
|
Fkbp11
|
FK506 binding protein 11 |
| chr3_+_59989282 | 6.13 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr12_-_58315949 | 5.64 |
ENSMUST00000062254.4
|
Clec14a
|
C-type lectin domain family 14, member a |
| chr2_-_147888816 | 5.52 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr16_-_45664664 | 5.42 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr19_-_58443830 | 5.41 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_+_12610668 | 5.35 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr18_-_56695333 | 5.17 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr19_-_58443593 | 4.90 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr6_+_129157576 | 4.71 |
ENSMUST00000032260.6
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr5_+_52521133 | 4.07 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr9_-_21671571 | 3.75 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr10_-_25412010 | 3.73 |
ENSMUST00000179685.3
|
Smlr1
|
small leucine-rich protein 1 |
| chr4_-_131776368 | 3.66 |
ENSMUST00000105981.9
ENSMUST00000084253.10 ENSMUST00000141291.3 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr7_-_3918484 | 3.10 |
ENSMUST00000038176.15
ENSMUST00000206077.2 ENSMUST00000090689.5 |
Lilra6
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 |
| chr7_-_3828640 | 3.05 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chrM_+_10167 | 2.89 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr17_+_25992761 | 2.67 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chrX_+_72830607 | 2.62 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr13_-_95754987 | 2.60 |
ENSMUST00000059193.7
|
F2r
|
coagulation factor II (thrombin) receptor |
| chr5_+_100054110 | 2.49 |
ENSMUST00000198837.3
|
Vamp9
|
vesicle-associated membrane protein 9 |
| chr11_-_115212851 | 2.47 |
ENSMUST00000103037.5
|
Ush1g
|
USH1 protein network component sans |
| chrM_+_9870 | 2.42 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr5_+_34683141 | 2.30 |
ENSMUST00000125817.8
ENSMUST00000067638.14 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr19_+_12824046 | 2.28 |
ENSMUST00000189517.2
|
Gm5244
|
predicted pseudogene 5244 |
| chr1_-_39844467 | 2.24 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr1_-_72251466 | 2.24 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr11_-_101062111 | 2.22 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr14_+_66378382 | 2.20 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr5_-_38649291 | 2.16 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr17_-_7652863 | 2.06 |
ENSMUST00000070059.5
|
Unc93a2
|
unc-93 homolog A2 |
| chr7_-_102540089 | 2.03 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
| chr10_-_81262948 | 2.03 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr8_+_105951777 | 1.92 |
ENSMUST00000034361.10
|
D230025D16Rik
|
RIKEN cDNA D230025D16 gene |
| chr14_-_123150497 | 1.83 |
ENSMUST00000162164.2
ENSMUST00000110679.9 ENSMUST00000161322.3 ENSMUST00000038075.12 |
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr17_+_7246289 | 1.83 |
ENSMUST00000179728.2
|
Rnaset2b
|
ribonuclease T2B |
| chr14_+_31888061 | 1.79 |
ENSMUST00000164341.2
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr14_+_65903878 | 1.75 |
ENSMUST00000069226.7
|
Scara5
|
scavenger receptor class A, member 5 |
| chr3_+_36530081 | 1.75 |
ENSMUST00000029268.7
|
1810062G17Rik
|
RIKEN cDNA 1810062G17 gene |
| chr14_-_55822696 | 1.75 |
ENSMUST00000022828.9
|
Emc9
|
ER membrane protein complex subunit 9 |
| chr9_+_39853511 | 1.74 |
ENSMUST00000062833.6
|
Olfr974
|
olfactory receptor 974 |
| chr5_+_45650821 | 1.72 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr17_+_46957151 | 1.65 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr17_+_37253802 | 1.62 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr14_+_65903840 | 1.60 |
ENSMUST00000022610.15
|
Scara5
|
scavenger receptor class A, member 5 |
| chr6_+_41279199 | 1.55 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr14_-_63654478 | 1.50 |
ENSMUST00000014597.5
|
Blk
|
B lymphoid kinase |
| chr4_-_107540726 | 1.46 |
ENSMUST00000131776.8
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr13_+_55875158 | 1.45 |
ENSMUST00000021958.6
ENSMUST00000124968.8 |
Pcbd2
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr17_+_7246365 | 1.44 |
ENSMUST00000232245.2
|
Rnaset2b
|
ribonuclease T2B |
| chr17_+_56935118 | 1.43 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chrX_+_30768610 | 1.38 |
ENSMUST00000179532.2
|
Btbd35f29
|
BTB domain containing 35, family member 29 |
| chr7_-_89914610 | 1.36 |
ENSMUST00000107221.9
ENSMUST00000107220.8 ENSMUST00000040413.2 |
Ccdc83
|
coiled-coil domain containing 83 |
| chr15_+_78210190 | 1.35 |
ENSMUST00000229034.2
ENSMUST00000096355.4 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr2_+_153716958 | 1.35 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr6_-_119444157 | 1.33 |
ENSMUST00000118120.8
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chrX_+_55939732 | 1.33 |
ENSMUST00000136396.8
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr8_+_3715747 | 1.29 |
ENSMUST00000014118.4
|
Mcemp1
|
mast cell expressed membrane protein 1 |
| chrX_+_3942699 | 1.25 |
ENSMUST00000140300.2
|
Btbd35f3
|
BTB domain containing 35, family member 3 |
| chr7_+_143383814 | 1.24 |
ENSMUST00000141916.8
ENSMUST00000144034.8 ENSMUST00000143338.2 ENSMUST00000207143.2 ENSMUST00000125564.2 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr1_-_138547879 | 1.21 |
ENSMUST00000187407.7
|
Nek7
|
NIMA (never in mitosis gene a)-related expressed kinase 7 |
| chrX_+_32750842 | 1.19 |
ENSMUST00000178827.3
|
Btbd35f12
|
BTB domain containing 35, family member 12 |
| chrX_+_4273978 | 1.18 |
ENSMUST00000105014.2
|
Btbd35f17
|
BTB domain containing 35, family member 17 |
| chrX_+_32411401 | 1.17 |
ENSMUST00000178747.3
|
Btbd35f5
|
BTB domain containing 35, family member 5 |
| chrX_-_55867668 | 1.17 |
ENSMUST00000135542.2
ENSMUST00000114766.8 |
Map7d3
|
MAP7 domain containing 3 |
| chr1_-_59134042 | 1.17 |
ENSMUST00000238601.2
ENSMUST00000238949.2 ENSMUST00000097080.4 |
C2cd6
|
C2 calcium dependent domain containing 6 |
| chr11_+_58531220 | 1.17 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58 |
| chrX_+_4855477 | 1.16 |
ENSMUST00000178143.2
|
Btbd35f27
|
BTB domain containing 35, family member 27 |
| chrX_+_4703838 | 1.16 |
ENSMUST00000105011.4
|
Btbd35f4
|
BTB domain containing 35, family member 4 |
| chr13_+_3588063 | 1.16 |
ENSMUST00000223396.2
ENSMUST00000059515.8 ENSMUST00000222365.2 |
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2 |
| chrX_-_33139812 | 1.15 |
ENSMUST00000105117.3
|
Btbd35f14
|
BTB domain containing 35, family member 14 |
| chrX_-_33014777 | 1.14 |
ENSMUST00000186329.2
|
Btbd35f15
|
BTB domain containing 35, family member 15 |
| chrX_+_32070863 | 1.14 |
ENSMUST00000238237.2
|
Btbd35f1
|
BTB domain containing 35, family member 1 |
| chrX_-_31034822 | 1.12 |
ENSMUST00000238426.2
|
Btbd35f19
|
BTB domain containing 35, family member 19 |
| chr2_-_76812799 | 1.11 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr2_+_36575800 | 1.10 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chrX_+_3605258 | 1.10 |
ENSMUST00000105019.3
|
Btbd35f18
|
BTB domain containing 35, family member 18 |
| chrX_-_3862027 | 1.09 |
ENSMUST00000185755.2
|
Btbd35f16
|
BTB domain containing 35, family member 16 |
| chrX_-_3657910 | 1.07 |
ENSMUST00000178621.2
|
Btbd35f10
|
BTB domain containing 35, family member 10 |
| chr1_+_159059869 | 1.05 |
ENSMUST00000076894.11
|
Cop1
|
COP1, E3 ubiquitin ligase |
| chr12_-_113771372 | 1.05 |
ENSMUST00000103456.4
|
Ighv2-7
|
immunoglobulin heavy variable 2-7 |
| chr11_+_101358990 | 1.02 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chrX_-_3398715 | 1.02 |
ENSMUST00000105020.2
|
Btbd35f11
|
BTB domain containing 35, family member 11 |
| chr17_-_8366536 | 1.01 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chrX_+_33094635 | 1.00 |
ENSMUST00000177912.2
|
Btbd35f13
|
BTB domain containing 35, family member 13 |
| chrX_+_11178173 | 0.99 |
ENSMUST00000178979.2
|
H2al1e
|
H2A histone family member L1E |
| chrX_+_31839202 | 0.99 |
ENSMUST00000179991.3
|
Btbd35f2
|
BTB domain containing 35, family member 2 |
| chr4_+_155646807 | 0.98 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chrX_-_3150852 | 0.95 |
ENSMUST00000178080.3
|
Btbd35f24
|
BTB domain containing 35, family member 24 |
| chr3_-_105839980 | 0.95 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr4_-_127224591 | 0.94 |
ENSMUST00000046532.4
|
Gjb3
|
gap junction protein, beta 3 |
| chrX_-_31375374 | 0.92 |
ENSMUST00000238772.2
|
Btbd35f25
|
BTB domain containing 35, family member 25 |
| chr17_+_37253916 | 0.92 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr19_+_34268053 | 0.90 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr5_-_46013838 | 0.88 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr14_+_20753074 | 0.87 |
ENSMUST00000071215.5
ENSMUST00000224633.2 |
Chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr7_-_102363618 | 0.87 |
ENSMUST00000084817.3
|
Olfr33
|
olfactory receptor 33 |
| chrX_+_3076875 | 0.86 |
ENSMUST00000189190.2
|
Btbd35f23
|
BTB domain containing 35, family member 23 |
| chrX_-_100865583 | 0.83 |
ENSMUST00000239206.2
|
Gm3858
|
predicted gene 3858 |
| chrX_+_4101601 | 0.83 |
ENSMUST00000105015.3
|
Btbd35f28
|
BTB domain containing 35, family member 28 |
| chrX_-_33394003 | 0.83 |
ENSMUST00000179466.2
|
Btbd35f6
|
BTB domain containing 35, family member 6 |
| chr15_+_31602252 | 0.79 |
ENSMUST00000042702.7
ENSMUST00000161061.3 |
Atpsckmt
|
ATP synthase C subunit lysine N-methyltransferase |
| chr7_+_17706049 | 0.78 |
ENSMUST00000094799.3
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr9_+_69902697 | 0.77 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr17_+_24161342 | 0.75 |
ENSMUST00000024940.11
ENSMUST00000182519.2 |
Sbp
|
spermine binding protein |
| chr19_-_10897993 | 0.74 |
ENSMUST00000025641.2
|
Zp1
|
zona pellucida glycoprotein 1 |
| chr1_+_161322219 | 0.73 |
ENSMUST00000086084.2
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr7_-_126613707 | 0.73 |
ENSMUST00000165096.9
|
Mvp
|
major vault protein |
| chr4_+_143341573 | 0.72 |
ENSMUST00000105773.2
|
Pramel21
|
PRAME like 21 |
| chr6_+_86172196 | 0.67 |
ENSMUST00000032066.13
|
Tgfa
|
transforming growth factor alpha |
| chr5_-_143123955 | 0.67 |
ENSMUST00000218872.3
|
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr6_+_56933451 | 0.66 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr4_+_114020656 | 0.65 |
ENSMUST00000145797.8
ENSMUST00000151810.2 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chrX_+_5581121 | 0.65 |
ENSMUST00000105007.3
|
Btbd35f7
|
BTB domain containing 35, family member 7 |
| chrX_-_33580888 | 0.64 |
ENSMUST00000238632.2
|
Btbd35f9
|
BTB domain containing 35, family member 9 |
| chr17_-_79190002 | 0.64 |
ENSMUST00000024884.5
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr19_+_34268071 | 0.62 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr5_-_129781323 | 0.61 |
ENSMUST00000042266.13
|
Septin14
|
septin 14 |
| chrX_+_31608531 | 0.59 |
ENSMUST00000238528.2
|
Btbd35f22
|
BTB domain containing 35, family member 22 |
| chr15_+_39997761 | 0.57 |
ENSMUST00000228780.2
|
9330182O14Rik
|
RIKEN cDNA 9330182O14 gene |
| chr7_-_110443557 | 0.56 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr17_+_25992742 | 0.52 |
ENSMUST00000134108.8
ENSMUST00000002350.11 |
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr3_-_63391300 | 0.50 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr14_-_56358080 | 0.50 |
ENSMUST00000089549.8
|
Gzme
|
granzyme E |
| chr6_+_149226891 | 0.49 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chr11_-_113540867 | 0.49 |
ENSMUST00000136392.8
ENSMUST00000125890.8 ENSMUST00000146031.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr4_-_84464521 | 0.49 |
ENSMUST00000177040.2
|
Bnc2
|
basonuclin 2 |
| chr7_-_139953579 | 0.48 |
ENSMUST00000074177.3
|
Olfr530
|
olfactory receptor 530 |
| chr4_-_144000597 | 0.48 |
ENSMUST00000105752.4
|
Pramel5
|
PRAME like 5 |
| chr10_-_85847697 | 0.48 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr1_-_69726384 | 0.48 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chrX_+_11184495 | 0.47 |
ENSMUST00000179859.2
|
H2al1g
|
H2A histone family member L1G |
| chr6_+_24857967 | 0.46 |
ENSMUST00000200968.4
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr7_+_17447163 | 0.44 |
ENSMUST00000081907.8
|
Ceacam5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr6_+_122967309 | 0.43 |
ENSMUST00000079379.3
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr1_-_161615927 | 0.42 |
ENSMUST00000193648.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr8_+_122677231 | 0.39 |
ENSMUST00000093078.13
ENSMUST00000170857.8 ENSMUST00000026354.15 ENSMUST00000174753.8 ENSMUST00000172511.8 |
Banp
|
BTG3 associated nuclear protein |
| chr7_-_24145107 | 0.39 |
ENSMUST00000205776.2
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr9_+_38026988 | 0.39 |
ENSMUST00000072290.5
|
Olfr889
|
olfactory receptor 889 |
| chr7_+_12661337 | 0.39 |
ENSMUST00000045870.5
|
Rnf225
|
ring finger protein 225 |
| chr4_+_114020581 | 0.38 |
ENSMUST00000079915.10
ENSMUST00000164297.8 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chr6_+_133269179 | 0.38 |
ENSMUST00000048459.8
|
5530400C23Rik
|
RIKEN cDNA 5530400C23 gene |
| chr2_+_89708781 | 0.37 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr9_-_54100726 | 0.37 |
ENSMUST00000034811.8
ENSMUST00000215736.2 |
Cyp19a1
|
cytochrome P450, family 19, subfamily a, polypeptide 1 |
| chr2_-_174909556 | 0.36 |
ENSMUST00000072895.10
ENSMUST00000109066.2 |
Gm14393
|
predicted gene 14393 |
| chr1_+_92547603 | 0.36 |
ENSMUST00000081274.6
|
Olfr12
|
olfactory receptor 12 |
| chr6_+_128911336 | 0.35 |
ENSMUST00000000254.14
|
Clec2g
|
C-type lectin domain family 2, member g |
| chrX_-_4194587 | 0.35 |
ENSMUST00000179325.2
|
Btbd35f20
|
BTB domain containing 35, family member 20 |
| chr13_-_26954110 | 0.34 |
ENSMUST00000055915.6
|
Hdgfl1
|
HDGF like 1 |
| chr11_+_97917520 | 0.33 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr11_+_74150634 | 0.32 |
ENSMUST00000217016.2
ENSMUST00000214303.2 |
Olfr406
|
olfactory receptor 406 |
| chr6_-_54570124 | 0.32 |
ENSMUST00000046520.13
|
Fkbp14
|
FK506 binding protein 14 |
| chr4_-_21767116 | 0.31 |
ENSMUST00000029915.6
|
Tstd3
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 3 |
| chr15_+_78210242 | 0.27 |
ENSMUST00000229678.2
ENSMUST00000231888.2 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr12_-_115567853 | 0.27 |
ENSMUST00000103538.3
ENSMUST00000198646.2 |
Ighv1-67
|
immunoglobulin heavy variable V1-67 |
| chr1_+_34537450 | 0.26 |
ENSMUST00000027299.10
|
Prss39
|
protease, serine 39 |
| chrX_+_32236876 | 0.26 |
ENSMUST00000105017.5
|
Btbd35f21
|
BTB domain containing 35, family member 21 |
| chr7_+_48608800 | 0.25 |
ENSMUST00000183659.8
|
Nav2
|
neuron navigator 2 |
| chr1_-_161616031 | 0.24 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr9_-_25063068 | 0.23 |
ENSMUST00000008573.9
|
Herpud2
|
HERPUD family member 2 |
| chr16_-_30406355 | 0.22 |
ENSMUST00000117363.9
|
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chrX_+_30987337 | 0.22 |
ENSMUST00000238765.2
|
Btbd35f26
|
BTB domain containing 35, family member 26 |
| chrX_+_14077387 | 0.22 |
ENSMUST00000105137.4
|
H2al1n
|
H2A histone family member L1N |
| chr17_-_33435325 | 0.21 |
ENSMUST00000112162.4
|
Olfr1564
|
olfactory receptor 1564 |
| chr7_-_33832640 | 0.20 |
ENSMUST00000038537.9
|
Wtip
|
WT1-interacting protein |
| chr7_+_19833605 | 0.19 |
ENSMUST00000165330.3
|
Vmn1r91
|
vomeronasal 1 receptor 91 |
| chr11_+_97917746 | 0.19 |
ENSMUST00000017548.7
|
Rpl19
|
ribosomal protein L19 |
| chr11_+_101333115 | 0.19 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr8_+_88290469 | 0.18 |
ENSMUST00000093342.6
|
4933402J07Rik
|
RIKEN cDNA 4933402J07 gene |
| chr4_-_45320579 | 0.17 |
ENSMUST00000030003.10
|
Exosc3
|
exosome component 3 |
| chr11_+_101333238 | 0.17 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr17_-_35246886 | 0.16 |
ENSMUST00000040151.9
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr6_-_40877303 | 0.14 |
ENSMUST00000063523.5
|
Prss58
|
protease, serine 58 |
| chr15_+_6552270 | 0.11 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr10_+_81164552 | 0.08 |
ENSMUST00000105325.4
ENSMUST00000220312.2 |
Tbxa2r
|
thromboxane A2 receptor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 4.2 | 12.5 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 3.9 | 19.6 | GO:0090467 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 3.7 | 15.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.6 | 13.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 2.6 | 20.9 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.8 | 5.5 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.7 | 11.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.3 | 10.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.2 | 8.7 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 1.1 | 13.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.9 | 2.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.6 | 4.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.5 | 5.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.5 | 1.5 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.4 | 14.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 14.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 1.2 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.3 | 8.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 0.7 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 6.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 1.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.2 | 6.3 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 3.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 7.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 2.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 2.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 4.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 3.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.2 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 5.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.6 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 2.8 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.1 | 2.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 2.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 11.0 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 1.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 1.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.7 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 5.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 2.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 7.8 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.9 | 20.9 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 3.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 2.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 10.5 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.3 | 5.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.6 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.2 | 1.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 1.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 2.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 19.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 9.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 11.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 5.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 10.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 17.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 9.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 10.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 4.9 | 14.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 4.3 | 13.0 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 3.9 | 11.8 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 3.0 | 20.9 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 2.1 | 16.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.8 | 12.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.8 | 19.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.5 | 13.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 1.3 | 5.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.3 | 5.2 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.9 | 9.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 4.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.7 | 3.4 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.7 | 2.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 9.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.6 | 5.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.5 | 9.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 1.5 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.4 | 15.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 1.2 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.4 | 1.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 6.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 1.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 1.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.3 | 5.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 5.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 2.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 5.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 16.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 6.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 10.5 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 10.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 4.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 1.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 2.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 19.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 11.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 10.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 7.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 14.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 7.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 16.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.4 | 15.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 9.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 10.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 5.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 37.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 6.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 12.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 4.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 3.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |