Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
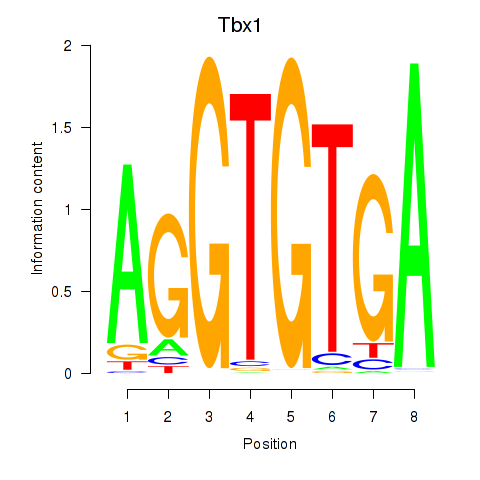
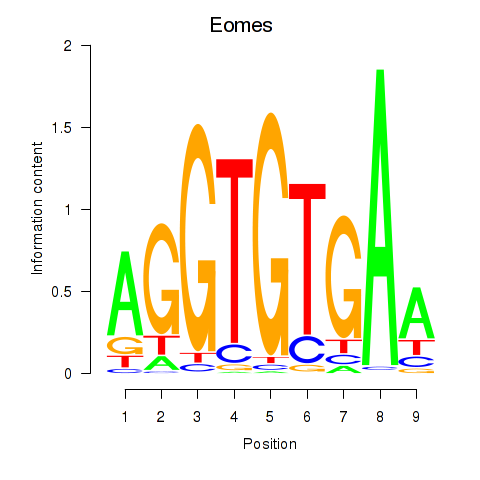
Results for Tbx1_Eomes
Z-value: 0.74
Transcription factors associated with Tbx1_Eomes
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx1
|
ENSMUSG00000009097.11 | Tbx1 |
|
Eomes
|
ENSMUSG00000032446.15 | Eomes |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx1 | mm39_v1_chr16_-_18405709_18405720 | 0.36 | 2.1e-03 | Click! |
| Eomes | mm39_v1_chr9_+_118307918_118307934 | -0.03 | 8.0e-01 | Click! |
Activity profile of Tbx1_Eomes motif
Sorted Z-values of Tbx1_Eomes motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx1_Eomes
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_100307601 | 6.69 |
ENSMUST00000138830.2
ENSMUST00000107044.10 ENSMUST00000116287.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr7_-_100307571 | 5.80 |
ENSMUST00000107043.8
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr2_-_57004933 | 5.21 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr6_-_127746390 | 5.20 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr9_-_99592116 | 5.10 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr4_+_42950367 | 4.96 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr2_-_13016570 | 4.91 |
ENSMUST00000061545.7
|
C1ql3
|
C1q-like 3 |
| chr9_-_15834052 | 4.66 |
ENSMUST00000217187.2
|
Fat3
|
FAT atypical cadherin 3 |
| chr11_-_83959175 | 4.38 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr6_+_114108190 | 4.36 |
ENSMUST00000032451.9
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
| chr12_+_88689638 | 3.92 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr11_+_53410552 | 3.67 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr18_-_34505544 | 3.58 |
ENSMUST00000236887.2
|
Reep5
|
receptor accessory protein 5 |
| chr6_-_116693849 | 3.54 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr3_-_158267771 | 3.48 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr4_+_125384481 | 3.46 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr18_+_61096660 | 3.38 |
ENSMUST00000039904.7
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr18_+_61096597 | 3.26 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr16_-_9812787 | 3.05 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr17_-_35954573 | 3.02 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr13_-_43457626 | 3.01 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr2_+_32498997 | 2.99 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr5_-_24806960 | 2.91 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr6_-_94677118 | 2.89 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr7_+_18618605 | 2.81 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr7_+_130179063 | 2.74 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_117706390 | 2.74 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_-_99592058 | 2.59 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr6_+_115111860 | 2.59 |
ENSMUST00000169345.4
|
Syn2
|
synapsin II |
| chr3_+_55369288 | 2.58 |
ENSMUST00000198412.5
ENSMUST00000199169.5 ENSMUST00000199702.5 ENSMUST00000198437.5 |
Dclk1
|
doublecortin-like kinase 1 |
| chrX_+_158623460 | 2.58 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_+_75464837 | 2.49 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chrX_+_40490005 | 2.46 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr9_+_50662625 | 2.39 |
ENSMUST00000217475.2
|
Cryab
|
crystallin, alpha B |
| chr17_-_31363245 | 2.33 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr2_+_54326329 | 2.30 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr4_+_43641262 | 2.28 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr3_+_102641822 | 2.25 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr6_+_115111872 | 2.18 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chr10_+_56253418 | 2.17 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr6_+_42263609 | 2.16 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr3_+_55369149 | 2.15 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr19_-_37184692 | 2.12 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr4_+_117706559 | 2.08 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_-_24956296 | 2.07 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr8_-_106660470 | 2.02 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr15_+_103411461 | 1.99 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr2_+_138120401 | 1.98 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr8_+_46338557 | 1.98 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chrX_+_162691978 | 1.96 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr9_+_27210500 | 1.93 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr8_+_46338498 | 1.89 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr11_+_98632953 | 1.84 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr1_-_134260666 | 1.84 |
ENSMUST00000168515.8
ENSMUST00000189361.2 |
Ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr11_+_98632696 | 1.83 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr18_-_60724855 | 1.83 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr9_+_75213570 | 1.81 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chrX_+_40490353 | 1.79 |
ENSMUST00000165288.2
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr3_+_55369384 | 1.78 |
ENSMUST00000200352.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr6_-_145811274 | 1.76 |
ENSMUST00000032386.11
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr1_+_66507523 | 1.73 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chrX_+_7786061 | 1.70 |
ENSMUST00000041096.4
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr16_-_9812410 | 1.67 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr6_+_7555053 | 1.67 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr6_-_145811028 | 1.63 |
ENSMUST00000111703.2
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr1_+_40619215 | 1.61 |
ENSMUST00000027233.9
|
Slc9a4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4 |
| chr4_-_49845549 | 1.58 |
ENSMUST00000093859.11
ENSMUST00000076674.4 |
Grin3a
|
glutamate receptor ionotropic, NMDA3A |
| chr11_+_98632631 | 1.57 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr16_+_62635039 | 1.56 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr13_-_89890609 | 1.50 |
ENSMUST00000109546.9
|
Vcan
|
versican |
| chr8_+_55024446 | 1.47 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr11_+_53410697 | 1.45 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chrM_+_7779 | 1.39 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr1_-_174749379 | 1.38 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr12_-_31684588 | 1.37 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr15_+_103411689 | 1.35 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chrX_-_58613428 | 1.32 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr3_-_30194559 | 1.30 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr15_+_74388044 | 1.29 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chrX_+_162692126 | 1.28 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr15_-_79897404 | 1.26 |
ENSMUST00000229912.2
ENSMUST00000229795.2 |
Pdgfb
|
platelet derived growth factor, B polypeptide |
| chrM_+_7758 | 1.24 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr1_+_153541412 | 1.23 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr1_+_153541020 | 1.23 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr12_+_69954506 | 1.22 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr6_+_72281587 | 1.21 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr7_+_25005510 | 1.21 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chrX_+_132809166 | 1.19 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr13_+_93440572 | 1.18 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr2_+_20524587 | 1.17 |
ENSMUST00000114604.9
ENSMUST00000066509.10 |
Etl4
|
enhancer trap locus 4 |
| chr6_+_15196950 | 1.15 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr6_+_42263644 | 1.15 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr4_+_84802513 | 1.13 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein |
| chr12_+_88920169 | 1.11 |
ENSMUST00000057634.14
|
Nrxn3
|
neurexin III |
| chr9_-_49710190 | 1.11 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr4_+_148085179 | 1.08 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr16_+_37597235 | 1.07 |
ENSMUST00000114763.3
|
Fstl1
|
follistatin-like 1 |
| chr19_-_10282218 | 1.07 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr8_+_124124493 | 1.02 |
ENSMUST00000212880.2
|
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr16_+_56298228 | 1.02 |
ENSMUST00000231832.2
ENSMUST00000096013.11 ENSMUST00000048471.15 ENSMUST00000231870.2 ENSMUST00000171000.3 ENSMUST00000231781.2 ENSMUST00000096012.11 |
Abi3bp
|
ABI family member 3 binding protein |
| chr10_+_116013256 | 1.02 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr18_-_65527078 | 0.99 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr11_+_121128042 | 0.96 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor |
| chr14_+_47121487 | 0.91 |
ENSMUST00000137543.9
|
Samd4
|
sterile alpha motif domain containing 4 |
| chr1_+_59563453 | 0.90 |
ENSMUST00000186434.3
|
Gm973
|
predicted gene 973 |
| chr9_-_49710058 | 0.88 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr10_+_80765900 | 0.87 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr1_-_151965876 | 0.86 |
ENSMUST00000044581.14
|
1700025G04Rik
|
RIKEN cDNA 1700025G04 gene |
| chrX_-_132799041 | 0.84 |
ENSMUST00000176718.8
ENSMUST00000176641.2 |
Tspan6
|
tetraspanin 6 |
| chrM_+_8603 | 0.83 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_+_67061908 | 0.83 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr15_-_11905697 | 0.81 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr18_+_69654572 | 0.81 |
ENSMUST00000200862.4
|
Tcf4
|
transcription factor 4 |
| chr10_-_35587888 | 0.80 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr12_+_3941728 | 0.80 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr11_+_67061837 | 0.80 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr18_+_69654900 | 0.79 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr10_+_116013122 | 0.78 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr8_+_120173458 | 0.78 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr10_+_103203552 | 0.77 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chr4_+_116078787 | 0.75 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr4_+_116078830 | 0.74 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr9_-_20984790 | 0.74 |
ENSMUST00000010348.7
|
Fdx2
|
ferredoxin 2 |
| chr11_-_102992434 | 0.73 |
ENSMUST00000103077.2
|
Plcd3
|
phospholipase C, delta 3 |
| chr4_-_35845204 | 0.72 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr7_-_81216687 | 0.71 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr14_-_70449438 | 0.69 |
ENSMUST00000227929.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr4_+_48045143 | 0.69 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr4_+_116078874 | 0.68 |
ENSMUST00000106490.3
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr1_-_87322443 | 0.68 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr1_+_63312420 | 0.66 |
ENSMUST00000239483.2
ENSMUST00000114132.8 ENSMUST00000126932.2 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr1_+_153541339 | 0.66 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr7_+_24967094 | 0.64 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chrX_+_55500170 | 0.63 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_-_108529375 | 0.60 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr19_-_18978463 | 0.60 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr5_-_24534554 | 0.60 |
ENSMUST00000115098.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr18_+_34973605 | 0.59 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr4_-_82423944 | 0.59 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr9_+_37119472 | 0.59 |
ENSMUST00000034632.10
|
Tmem218
|
transmembrane protein 218 |
| chr16_-_16345197 | 0.58 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_-_56392646 | 0.58 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr16_-_32688640 | 0.57 |
ENSMUST00000089684.10
ENSMUST00000040986.15 ENSMUST00000115105.9 |
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr5_-_135518098 | 0.57 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr3_+_96537235 | 0.56 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr5_-_148336711 | 0.56 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr9_+_86897590 | 0.55 |
ENSMUST00000058846.11
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr3_-_94490023 | 0.55 |
ENSMUST00000029783.16
|
Snx27
|
sorting nexin family member 27 |
| chrX_-_58612709 | 0.54 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr7_-_102804952 | 0.53 |
ENSMUST00000061055.2
|
Olfr589
|
olfactory receptor 589 |
| chr4_+_53440389 | 0.53 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chrX_+_41591355 | 0.53 |
ENSMUST00000189753.7
|
Sh2d1a
|
SH2 domain containing 1A |
| chr18_+_69654992 | 0.51 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr2_+_25162487 | 0.51 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr5_+_111881790 | 0.49 |
ENSMUST00000180627.2
|
Gm26897
|
predicted gene, 26897 |
| chr1_-_69147185 | 0.47 |
ENSMUST00000121473.8
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr2_-_147888816 | 0.47 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chrM_+_9459 | 0.47 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr17_+_49239393 | 0.47 |
ENSMUST00000046254.3
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr6_-_92191823 | 0.47 |
ENSMUST00000203516.2
|
Rbsn
|
rabenosyn, RAB effector |
| chr19_+_22670134 | 0.46 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr4_+_84802592 | 0.46 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr9_+_71123061 | 0.46 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr4_+_84802650 | 0.45 |
ENSMUST00000169371.9
|
Cntln
|
centlein, centrosomal protein |
| chr3_+_96537484 | 0.45 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr19_-_45619559 | 0.45 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr2_+_71359000 | 0.42 |
ENSMUST00000126400.2
|
Dlx1
|
distal-less homeobox 1 |
| chrX_+_150799414 | 0.42 |
ENSMUST00000045312.6
|
Smc1a
|
structural maintenance of chromosomes 1A |
| chr9_-_58109627 | 0.42 |
ENSMUST00000216231.2
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chrM_+_9870 | 0.42 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr9_+_110948492 | 0.41 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr7_+_107702486 | 0.40 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chr11_-_22236795 | 0.40 |
ENSMUST00000180360.8
ENSMUST00000109563.9 |
Ehbp1
|
EH domain binding protein 1 |
| chr9_-_110474398 | 0.39 |
ENSMUST00000149089.2
|
Nbeal2
|
neurobeachin-like 2 |
| chr17_-_57289121 | 0.38 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr5_-_24556602 | 0.38 |
ENSMUST00000036092.10
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr11_+_86574811 | 0.38 |
ENSMUST00000108022.8
ENSMUST00000108021.2 |
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr7_-_141698342 | 0.37 |
ENSMUST00000210537.2
|
Gm45337
|
predicted gene 45337 |
| chr3_+_104688363 | 0.37 |
ENSMUST00000002298.7
|
Ppm1j
|
protein phosphatase 1J |
| chr18_+_65715460 | 0.37 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chrX_-_100463395 | 0.35 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr14_+_30973407 | 0.34 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr17_-_81035453 | 0.34 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chrM_+_10167 | 0.34 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr6_-_92191878 | 0.33 |
ENSMUST00000014694.11
|
Rbsn
|
rabenosyn, RAB effector |
| chr11_+_49737558 | 0.33 |
ENSMUST00000109179.9
ENSMUST00000178543.8 ENSMUST00000164643.8 ENSMUST00000020634.14 |
Mapk9
|
mitogen-activated protein kinase 9 |
| chr9_-_107166543 | 0.30 |
ENSMUST00000192054.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr8_-_32408380 | 0.30 |
ENSMUST00000208497.3
ENSMUST00000207584.3 |
Nrg1
|
neuregulin 1 |
| chr2_-_25162347 | 0.30 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr10_+_115653152 | 0.29 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr7_-_98305986 | 0.29 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr11_-_51748450 | 0.28 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr2_+_163662752 | 0.27 |
ENSMUST00000029188.8
|
Ccn5
|
cellular communication network factor 5 |
| chr14_-_24294933 | 0.25 |
ENSMUST00000169880.3
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr9_+_15150341 | 0.25 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr15_+_99568208 | 0.23 |
ENSMUST00000023758.9
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr6_-_83504756 | 0.22 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_-_85752765 | 0.22 |
ENSMUST00000037646.9
ENSMUST00000220032.2 |
Prdm4
|
PR domain containing 4 |
| chrX_-_100463810 | 0.21 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr9_-_35122261 | 0.20 |
ENSMUST00000043805.15
ENSMUST00000142595.8 ENSMUST00000127996.8 |
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.3 | 5.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.3 | 5.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.1 | 7.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.0 | 5.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.0 | 2.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.9 | 2.8 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.7 | 2.2 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.7 | 2.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 4.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.6 | 1.7 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 2.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.5 | 6.6 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.5 | 1.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.5 | 1.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 1.3 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.4 | 2.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 3.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 2.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 1.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 2.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 1.1 | GO:0099542 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.3 | 1.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 0.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 0.8 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 1.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 3.3 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 | 3.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 3.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.7 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.2 | 2.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 3.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 3.4 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.2 | 4.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 5.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 6.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.6 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.4 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 1.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 0.5 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 2.7 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.8 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.8 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 3.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.8 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.4 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 0.6 | GO:0032324 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 1.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 2.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 2.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.4 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.6 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 3.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 4.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 1.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.8 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.3 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.1 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.3 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 2.2 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 3.2 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 7.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 5.6 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 4.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 2.1 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0040030 | nucleosome positioning(GO:0016584) regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 1.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 5.6 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.5 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 2.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.6 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.8 | 2.5 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.3 | 4.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 6.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 2.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 4.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 14.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 6.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.8 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 3.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 2.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 3.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 7.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 3.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 6.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 9.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 3.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 2.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 8.2 | GO:0036477 | somatodendritic compartment(GO:0036477) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.1 | 3.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.9 | 5.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.6 | 4.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 3.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 3.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.5 | 2.8 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 6.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 7.9 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.4 | 6.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 1.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 2.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 3.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 3.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 4.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.8 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.2 | 2.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 5.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 3.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 5.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 2.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 2.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 14.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 4.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 1.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 2.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 7.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 7.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 4.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 7.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 11.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.8 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 2.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 2.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 3.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |