Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
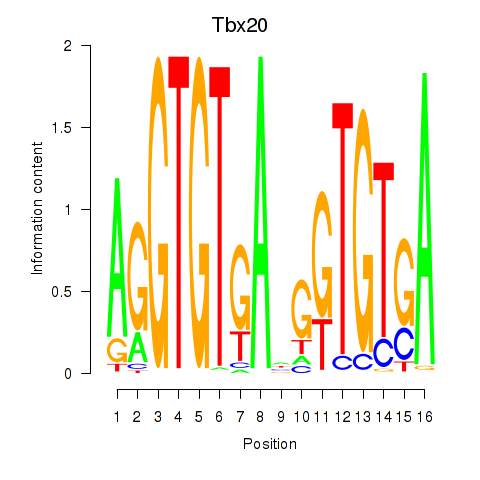
Results for Tbx20
Z-value: 0.49
Transcription factors associated with Tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx20
|
ENSMUSG00000031965.16 | Tbx20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx20 | mm39_v1_chr9_-_24685572_24685599 | -0.37 | 1.2e-03 | Click! |
Activity profile of Tbx20 motif
Sorted Z-values of Tbx20 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx20
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_122129379 | 4.56 |
ENSMUST00000028656.2
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr2_-_122128930 | 3.65 |
ENSMUST00000237546.2
|
Duox2
|
dual oxidase 2 |
| chr8_-_106660470 | 3.64 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr8_-_3744167 | 3.07 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr4_-_118294521 | 2.88 |
ENSMUST00000006565.13
|
Cdc20
|
cell division cycle 20 |
| chr19_+_38043446 | 2.41 |
ENSMUST00000236044.2
ENSMUST00000116506.8 ENSMUST00000096096.12 ENSMUST00000169673.3 |
Cep55
|
centrosomal protein 55 |
| chr9_-_99592116 | 2.12 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr7_+_79309938 | 2.10 |
ENSMUST00000035977.9
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr5_-_53864874 | 2.04 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr9_-_99592058 | 1.90 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr12_-_114487525 | 1.83 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr11_+_98239230 | 1.37 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr5_-_120915693 | 1.18 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr6_+_15196950 | 1.10 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr12_+_51395047 | 0.99 |
ENSMUST00000119211.8
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr12_+_51395154 | 0.98 |
ENSMUST00000121521.2
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr17_-_23964807 | 0.79 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr15_+_75140241 | 0.62 |
ENSMUST00000023247.13
|
Ly6f
|
lymphocyte antigen 6 complex, locus F |
| chr3_-_143908111 | 0.57 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr1_-_136876902 | 0.55 |
ENSMUST00000195428.2
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_-_143908060 | 0.53 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr2_+_74512638 | 0.51 |
ENSMUST00000142312.3
|
Hoxd11
|
homeobox D11 |
| chr15_-_76702170 | 0.48 |
ENSMUST00000175843.3
ENSMUST00000177026.3 ENSMUST00000176736.3 ENSMUST00000036176.16 ENSMUST00000176219.9 ENSMUST00000239134.2 ENSMUST00000239003.2 ENSMUST00000077821.10 |
Arhgap39
|
Rho GTPase activating protein 39 |
| chr8_+_129793635 | 0.44 |
ENSMUST00000026912.8
|
Ccdc7b
|
coiled-coil domain containing 7B |
| chr8_-_86281946 | 0.37 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr10_+_75160692 | 0.32 |
ENSMUST00000217703.2
|
Adora2a
|
adenosine A2a receptor |
| chr8_+_129793610 | 0.27 |
ENSMUST00000108745.8
|
Ccdc7b
|
coiled-coil domain containing 7B |
| chr11_-_23720953 | 0.24 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr16_-_19241884 | 0.23 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr1_-_136877277 | 0.21 |
ENSMUST00000168126.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr8_-_129791969 | 0.20 |
ENSMUST00000125112.8
ENSMUST00000108747.3 ENSMUST00000214889.2 ENSMUST00000095158.11 |
Ccdc7a
|
coiled-coil domain containing 7A |
| chr6_-_122317457 | 0.17 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chrY_+_7722941 | 0.12 |
ENSMUST00000189007.2
|
Gm28576
|
predicted gene 28576 |
| chr7_-_105436775 | 0.12 |
ENSMUST00000078482.13
|
Dchs1
|
dachsous cadherin related 1 |
| chr15_-_102425241 | 0.10 |
ENSMUST00000169162.8
ENSMUST00000023812.10 ENSMUST00000165174.8 ENSMUST00000169367.8 ENSMUST00000169377.8 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chrY_+_7253787 | 0.10 |
ENSMUST00000186194.2
|
Gm28998
|
predicted gene 28998 |
| chrX_-_133652080 | 0.08 |
ENSMUST00000113194.8
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr5_+_57879201 | 0.05 |
ENSMUST00000199310.2
|
Pcdh7
|
protocadherin 7 |
| chrX_-_133652140 | 0.03 |
ENSMUST00000052431.12
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr2_-_88157559 | 0.02 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr9_+_72946994 | 0.01 |
ENSMUST00000184126.3
|
Pigbos1
|
Pigb opposite strand 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.6 | 2.9 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.6 | 4.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 4.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.4 | 3.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.3 | 2.0 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 | 2.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 1.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 1.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 2.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 1.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.3 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.1 | 0.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 2.0 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 1.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 4.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.5 | 1.4 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.4 | 3.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 3.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 2.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 4.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |