Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
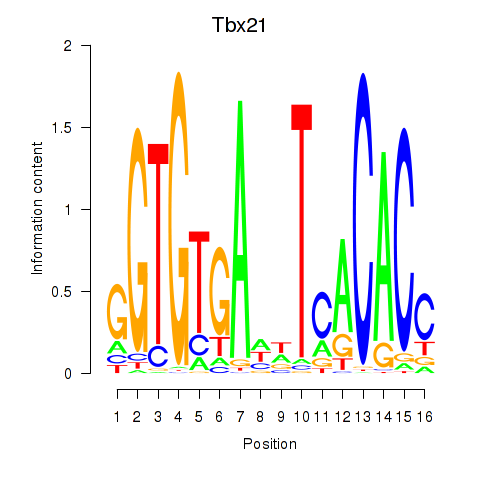
Results for Tbx21
Z-value: 1.08
Transcription factors associated with Tbx21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx21
|
ENSMUSG00000001444.3 | Tbx21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx21 | mm39_v1_chr11_-_97006153_97006158 | 0.48 | 2.2e-05 | Click! |
Activity profile of Tbx21 motif
Sorted Z-values of Tbx21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110248815 | 11.87 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr12_-_113223839 | 8.14 |
ENSMUST00000194738.6
ENSMUST00000178282.3 |
Igha
|
immunoglobulin heavy constant alpha |
| chr5_-_120915693 | 7.46 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr10_+_77366086 | 6.82 |
ENSMUST00000000299.14
ENSMUST00000131023.8 |
Itgb2
|
integrin beta 2 |
| chr2_-_164198427 | 6.76 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr6_+_129374260 | 6.59 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr17_+_48554786 | 6.33 |
ENSMUST00000048065.6
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr6_+_129374441 | 6.14 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr7_-_126641593 | 5.81 |
ENSMUST00000032915.8
|
Kif22
|
kinesin family member 22 |
| chr6_+_68414401 | 5.46 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr6_+_40868154 | 5.39 |
ENSMUST00000103262.3
|
Trbv1
|
T cell receptor beta, variable 1 |
| chr10_+_43455919 | 5.30 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen |
| chr16_-_58319696 | 5.06 |
ENSMUST00000148061.2
|
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr12_-_4924341 | 4.87 |
ENSMUST00000137337.8
ENSMUST00000045921.14 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr12_-_70274433 | 4.78 |
ENSMUST00000071250.13
|
Pygl
|
liver glycogen phosphorylase |
| chr1_+_74193138 | 4.75 |
ENSMUST00000027372.8
ENSMUST00000106899.4 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr6_-_60806810 | 4.59 |
ENSMUST00000163779.8
|
Snca
|
synuclein, alpha |
| chr10_+_77366195 | 4.52 |
ENSMUST00000130059.8
|
Itgb2
|
integrin beta 2 |
| chr17_+_33774681 | 4.39 |
ENSMUST00000087605.13
ENSMUST00000174695.2 |
Myo1f
|
myosin IF |
| chr6_-_70435020 | 3.86 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr12_-_113843161 | 3.80 |
ENSMUST00000103451.5
|
Ighv2-9
|
immunoglobulin heavy variable 2-9 |
| chr7_+_127661807 | 3.72 |
ENSMUST00000064821.14
|
Itgam
|
integrin alpha M |
| chr7_-_44775961 | 3.72 |
ENSMUST00000210931.3
|
Rpl13a
|
ribosomal protein L13A |
| chr15_+_98972850 | 3.62 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein |
| chr2_-_121273054 | 3.59 |
ENSMUST00000116432.2
|
Ell3
|
elongation factor RNA polymerase II-like 3 |
| chr17_+_36172210 | 3.57 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr1_+_133058881 | 3.48 |
ENSMUST00000052529.4
|
Ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr7_+_140658101 | 3.47 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr7_-_126641565 | 3.39 |
ENSMUST00000205806.2
|
Kif22
|
kinesin family member 22 |
| chr6_-_70237939 | 3.39 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr11_+_103061905 | 3.38 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr1_+_171667259 | 3.30 |
ENSMUST00000155802.9
|
Cd84
|
CD84 antigen |
| chr9_+_59198829 | 3.14 |
ENSMUST00000217570.2
ENSMUST00000026266.9 |
Adpgk
|
ADP-dependent glucokinase |
| chr7_+_127661835 | 3.14 |
ENSMUST00000106242.10
ENSMUST00000120355.8 ENSMUST00000106240.9 ENSMUST00000098015.10 |
Itgam
Gm49368
|
integrin alpha M predicted gene, 49368 |
| chr11_-_118139418 | 2.85 |
ENSMUST00000106305.9
|
Cyth1
|
cytohesin 1 |
| chr11_+_24030663 | 2.83 |
ENSMUST00000118955.2
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr15_+_79784365 | 2.78 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr9_+_69360902 | 2.77 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr11_-_118139331 | 2.77 |
ENSMUST00000017276.14
|
Cyth1
|
cytohesin 1 |
| chr9_+_50476637 | 2.64 |
ENSMUST00000214117.2
|
Il18
|
interleukin 18 |
| chr13_+_100806206 | 2.63 |
ENSMUST00000170347.4
ENSMUST00000057325.15 |
Ccdc125
|
coiled-coil domain containing 125 |
| chr7_+_79944198 | 2.62 |
ENSMUST00000163812.9
ENSMUST00000047558.14 ENSMUST00000174199.8 ENSMUST00000173824.8 ENSMUST00000174172.8 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr1_-_97589365 | 2.61 |
ENSMUST00000153115.8
ENSMUST00000142234.2 |
Macir
|
macrophage immunometabolism regulator |
| chr17_-_27247891 | 2.49 |
ENSMUST00000078691.12
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr19_+_38043506 | 2.35 |
ENSMUST00000237408.2
|
Cep55
|
centrosomal protein 55 |
| chr5_-_121045568 | 2.29 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr19_+_34169629 | 2.28 |
ENSMUST00000239240.2
ENSMUST00000054956.15 |
Stambpl1
|
STAM binding protein like 1 |
| chr8_-_57940834 | 2.21 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr19_+_12438125 | 2.18 |
ENSMUST00000081035.9
|
Mpeg1
|
macrophage expressed gene 1 |
| chr15_+_74586682 | 2.17 |
ENSMUST00000023265.5
|
Psca
|
prostate stem cell antigen |
| chr13_-_74956030 | 2.16 |
ENSMUST00000065629.6
|
Cast
|
calpastatin |
| chr9_+_44966464 | 2.11 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr12_+_116369017 | 2.10 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr12_-_70278188 | 2.08 |
ENSMUST00000161083.2
|
Pygl
|
liver glycogen phosphorylase |
| chr6_+_113355076 | 2.03 |
ENSMUST00000156898.5
ENSMUST00000203578.3 ENSMUST00000171058.8 |
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr7_-_119058489 | 2.03 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr3_-_54642450 | 1.94 |
ENSMUST00000153224.2
|
Exosc8
|
exosome component 8 |
| chr11_+_116421923 | 1.93 |
ENSMUST00000138840.8
|
Sphk1
|
sphingosine kinase 1 |
| chr19_-_5416339 | 1.92 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr9_+_59446823 | 1.89 |
ENSMUST00000026262.8
|
Hexa
|
hexosaminidase A |
| chr19_+_53298906 | 1.77 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr7_+_4340708 | 1.75 |
ENSMUST00000006792.6
ENSMUST00000126417.3 |
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr19_+_5416769 | 1.75 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr5_-_148988110 | 1.72 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr6_-_70412460 | 1.65 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr2_+_164674782 | 1.61 |
ENSMUST00000103093.10
|
Ctsa
|
cathepsin A |
| chr18_+_89215438 | 1.60 |
ENSMUST00000237110.2
|
Cd226
|
CD226 antigen |
| chr9_-_85631361 | 1.59 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr11_+_72326337 | 1.55 |
ENSMUST00000076443.10
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr11_+_72326391 | 1.54 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr15_+_54274151 | 1.52 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr5_+_21391282 | 1.51 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr19_-_5416626 | 1.50 |
ENSMUST00000237167.2
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr16_-_35951553 | 1.49 |
ENSMUST00000161638.2
ENSMUST00000096090.3 |
Csta1
|
cystatin A1 |
| chr4_-_117013396 | 1.48 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr18_+_67221287 | 1.48 |
ENSMUST00000025402.15
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr8_-_106427696 | 1.46 |
ENSMUST00000042608.8
|
Acd
|
adrenocortical dysplasia |
| chr11_+_70342731 | 1.43 |
ENSMUST00000151013.8
ENSMUST00000019067.8 |
Med11
|
mediator complex subunit 11 |
| chr13_+_13764982 | 1.42 |
ENSMUST00000110559.3
|
Lyst
|
lysosomal trafficking regulator |
| chr11_+_72326358 | 1.42 |
ENSMUST00000108499.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr6_-_128803182 | 1.41 |
ENSMUST00000204756.3
ENSMUST00000204394.3 ENSMUST00000204423.3 ENSMUST00000204677.2 ENSMUST00000205130.3 ENSMUST00000174544.2 ENSMUST00000172887.8 ENSMUST00000032472.11 |
Gm44511
Klrb1b
|
predicted gene 44511 killer cell lectin-like receptor subfamily B member 1B |
| chr8_+_85428391 | 1.39 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr8_+_89015705 | 1.38 |
ENSMUST00000171456.9
|
Adcy7
|
adenylate cyclase 7 |
| chr11_-_74480870 | 1.36 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr17_-_56428968 | 1.34 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr9_+_107926441 | 1.32 |
ENSMUST00000112295.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr7_+_35148188 | 1.29 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr14_+_53947359 | 1.26 |
ENSMUST00000103638.6
|
Trav6-7-dv9
|
T cell receptor alpha variable 6-7-DV9 |
| chr4_+_156214969 | 1.24 |
ENSMUST00000209248.2
|
Rnf223
|
ring finger 223 |
| chr11_+_59433508 | 1.23 |
ENSMUST00000101148.9
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr2_-_151815307 | 1.21 |
ENSMUST00000109863.2
|
Fam110a
|
family with sequence similarity 110, member A |
| chr1_-_16174387 | 1.20 |
ENSMUST00000149566.2
|
Rpl7
|
ribosomal protein L7 |
| chr2_-_23045876 | 1.20 |
ENSMUST00000028119.7
|
Mastl
|
microtubule associated serine/threonine kinase-like |
| chr11_-_70873773 | 1.19 |
ENSMUST00000078528.7
|
C1qbp
|
complement component 1, q subcomponent binding protein |
| chr7_+_35148579 | 1.19 |
ENSMUST00000032703.10
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr13_+_22129246 | 1.18 |
ENSMUST00000176511.8
ENSMUST00000102978.8 ENSMUST00000152258.9 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr15_-_34495329 | 1.15 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr11_+_120499295 | 1.12 |
ENSMUST00000106194.8
ENSMUST00000106195.3 ENSMUST00000061309.5 |
Npb
|
neuropeptide B |
| chr9_+_69361348 | 1.10 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr9_+_107926502 | 1.01 |
ENSMUST00000047947.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr2_+_164674801 | 0.94 |
ENSMUST00000103092.9
ENSMUST00000151493.3 ENSMUST00000127650.8 |
Ctsa
|
cathepsin A |
| chr1_+_59952131 | 0.93 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chrX_+_73298388 | 0.93 |
ENSMUST00000119197.8
ENSMUST00000088313.5 |
Emd
|
emerin |
| chr8_+_72704882 | 0.92 |
ENSMUST00000131237.8
ENSMUST00000136516.9 ENSMUST00000109997.10 ENSMUST00000132848.2 |
Zfp961
|
zinc finger protein 961 |
| chr12_+_73333641 | 0.91 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr3_-_92715195 | 0.89 |
ENSMUST00000179917.2
|
Lce1k
|
late cornified envelope 1K |
| chr5_-_138169253 | 0.85 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr3_-_15892807 | 0.82 |
ENSMUST00000148194.8
|
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr2_+_154975419 | 0.77 |
ENSMUST00000029126.15
ENSMUST00000109685.8 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr3_-_98247237 | 0.76 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr3_-_103698391 | 0.76 |
ENSMUST00000106845.9
ENSMUST00000029438.15 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr11_-_45845992 | 0.74 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr9_+_40059408 | 0.73 |
ENSMUST00000046333.9
ENSMUST00000238613.2 |
Tmem225
|
transmembrane protein 225 |
| chr7_+_35148461 | 0.72 |
ENSMUST00000118969.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr17_+_37977879 | 0.71 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr12_-_8589545 | 0.71 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr4_+_132366298 | 0.70 |
ENSMUST00000135299.8
ENSMUST00000020197.14 ENSMUST00000180250.8 ENSMUST00000081726.13 ENSMUST00000079157.11 |
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr17_+_43327412 | 0.66 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr6_-_50238723 | 0.65 |
ENSMUST00000101405.10
ENSMUST00000165099.8 ENSMUST00000170142.8 |
Gsdme
|
gasdermin E |
| chr15_+_80017315 | 0.64 |
ENSMUST00000023050.9
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr7_+_134870237 | 0.63 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr16_-_95928804 | 0.63 |
ENSMUST00000233292.2
ENSMUST00000050884.16 |
Hmgn1
|
high mobility group nucleosomal binding domain 1 |
| chr15_-_63707798 | 0.62 |
ENSMUST00000089900.10
|
Gsdmc2
|
gasdermin C2 |
| chr7_-_26878260 | 0.62 |
ENSMUST00000093040.13
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr11_-_71121559 | 0.61 |
ENSMUST00000108516.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr4_+_135673758 | 0.60 |
ENSMUST00000030432.8
|
Hmgcl
|
3-hydroxy-3-methylglutaryl-Coenzyme A lyase |
| chr2_-_151815759 | 0.59 |
ENSMUST00000109865.8
ENSMUST00000109864.8 |
Fam110a
|
family with sequence similarity 110, member A |
| chr10_+_75983285 | 0.58 |
ENSMUST00000020450.4
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr13_-_74956640 | 0.57 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr8_-_71349927 | 0.55 |
ENSMUST00000212709.2
ENSMUST00000212796.2 ENSMUST00000212378.2 ENSMUST00000054220.10 ENSMUST00000212494.2 |
Rpl18a
|
ribosomal protein L18A |
| chr15_+_34495441 | 0.53 |
ENSMUST00000052290.14
ENSMUST00000079028.6 |
Pop1
|
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
| chr7_-_141783708 | 0.52 |
ENSMUST00000097942.3
|
Krtap5-5
|
keratin associated protein 5-5 |
| chr10_+_7668655 | 0.52 |
ENSMUST00000015901.11
|
Ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr3_+_142236086 | 0.51 |
ENSMUST00000171263.8
ENSMUST00000045097.11 |
Gbp7
|
guanylate binding protein 7 |
| chr11_-_45846291 | 0.51 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr7_-_99477551 | 0.50 |
ENSMUST00000036331.7
|
Neu3
|
neuraminidase 3 |
| chr18_-_42712717 | 0.48 |
ENSMUST00000054738.5
|
Gpr151
|
G protein-coupled receptor 151 |
| chr16_+_22769822 | 0.46 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chrX_+_73298285 | 0.43 |
ENSMUST00000002029.13
|
Emd
|
emerin |
| chr16_+_22769844 | 0.43 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr2_+_122065230 | 0.42 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr1_+_88093726 | 0.38 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr9_+_38120348 | 0.38 |
ENSMUST00000093867.3
|
Olfr893
|
olfactory receptor 893 |
| chr13_-_100787481 | 0.38 |
ENSMUST00000177848.3
ENSMUST00000022136.14 |
Rad17
|
RAD17 checkpoint clamp loader component |
| chr13_-_27574822 | 0.37 |
ENSMUST00000102954.10
ENSMUST00000091678.3 |
Prl2b1
|
prolactin family 2, subfamily b, member 1 |
| chr19_+_5475149 | 0.37 |
ENSMUST00000159759.3
ENSMUST00000054477.8 |
AI837181
|
expressed sequence AI837181 |
| chr14_+_56255422 | 0.36 |
ENSMUST00000022836.6
|
Mcpt1
|
mast cell protease 1 |
| chr14_-_60488922 | 0.35 |
ENSMUST00000225311.2
ENSMUST00000041905.8 |
Gm49336
|
predicted gene, 49336 |
| chr17_-_24746804 | 0.34 |
ENSMUST00000176353.8
ENSMUST00000176237.8 |
Traf7
|
TNF receptor-associated factor 7 |
| chr14_-_51287620 | 0.33 |
ENSMUST00000076106.4
|
Rnase11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr7_-_126460820 | 0.32 |
ENSMUST00000129812.2
ENSMUST00000106342.8 |
Ino80e
|
INO80 complex subunit E |
| chr6_-_113354468 | 0.31 |
ENSMUST00000099118.8
|
Tada3
|
transcriptional adaptor 3 |
| chr12_+_86468556 | 0.30 |
ENSMUST00000021680.12
|
Esrrb
|
estrogen related receptor, beta |
| chr10_+_129610507 | 0.29 |
ENSMUST00000203598.3
|
Olfr809
|
olfactory receptor 809 |
| chr12_-_114217671 | 0.28 |
ENSMUST00000193408.2
|
Ighv3-4
|
immunoglobulin heavy variable V3-4 |
| chr19_-_60215097 | 0.28 |
ENSMUST00000238054.2
ENSMUST00000065286.2 |
Fam204a
|
family with sequence similarity 204, member A |
| chr13_+_21919225 | 0.27 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr12_-_31704766 | 0.27 |
ENSMUST00000020977.4
|
Dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr17_-_24746911 | 0.26 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7 |
| chr3_-_107145968 | 0.26 |
ENSMUST00000197758.5
|
Prok1
|
prokineticin 1 |
| chr10_+_127894843 | 0.26 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr17_+_39347585 | 0.25 |
ENSMUST00000232955.2
|
Esp24
|
exocrine gland secreted peptide 24 |
| chr11_+_59433554 | 0.24 |
ENSMUST00000149126.2
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr2_+_90152544 | 0.24 |
ENSMUST00000214797.3
|
Olfr1565
|
olfactory receptor 1565 |
| chr2_-_86857424 | 0.23 |
ENSMUST00000214857.2
ENSMUST00000215972.2 |
Olfr1104
|
olfactory receptor 1104 |
| chrY_+_12383184 | 0.23 |
ENSMUST00000179220.2
|
Gm20812
|
predicted gene, 20812 |
| chr18_-_3309858 | 0.23 |
ENSMUST00000144496.8
ENSMUST00000154715.8 |
Crem
|
cAMP responsive element modulator |
| chr7_+_43896146 | 0.22 |
ENSMUST00000055858.14
ENSMUST00000137702.8 ENSMUST00000188111.7 ENSMUST00000185481.7 ENSMUST00000146155.8 ENSMUST00000107949.8 ENSMUST00000084937.11 ENSMUST00000107950.9 ENSMUST00000107948.8 ENSMUST00000187524.7 ENSMUST00000137742.8 ENSMUST00000125318.3 |
2410002F23Rik
Gm28496
|
RIKEN cDNA 2410002F23 gene predicted gene 28496 |
| chr4_+_115420817 | 0.21 |
ENSMUST00000141033.8
ENSMUST00000030486.15 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr2_-_152382014 | 0.21 |
ENSMUST00000060598.4
|
Defb29
|
defensin beta 29 |
| chr5_-_90031180 | 0.21 |
ENSMUST00000198151.2
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 |
| chr4_-_118792037 | 0.20 |
ENSMUST00000081960.5
|
Olfr1328
|
olfactory receptor 1328 |
| chr2_-_166885414 | 0.17 |
ENSMUST00000067584.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr1_-_192812509 | 0.16 |
ENSMUST00000085555.7
|
Utp25
|
UTP25 small subunit processome component |
| chr4_+_151014167 | 0.15 |
ENSMUST00000126707.3
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr17_-_42922286 | 0.15 |
ENSMUST00000068355.8
|
Opn5
|
opsin 5 |
| chr4_-_132080916 | 0.15 |
ENSMUST00000155129.2
ENSMUST00000151374.2 |
Rcc1
Snhg3
|
regulator of chromosome condensation 1 small nucleolar RNA host gene 3 |
| chr14_-_40728839 | 0.14 |
ENSMUST00000152837.2
ENSMUST00000134715.8 |
Prxl2a
|
peroxiredoxin like 2A |
| chr2_+_90229027 | 0.14 |
ENSMUST00000216111.3
|
Olfr1274
|
olfactory receptor 1274 |
| chr6_+_15727798 | 0.14 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr7_-_43896112 | 0.13 |
ENSMUST00000237383.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chrX_+_73298342 | 0.12 |
ENSMUST00000096424.11
|
Emd
|
emerin |
| chr8_+_110835424 | 0.11 |
ENSMUST00000219454.2
|
Tle7
|
TLE family member 7 |
| chr9_+_65370077 | 0.11 |
ENSMUST00000215170.2
|
Spg21
|
SPG21, maspardin |
| chr14_+_103750763 | 0.11 |
ENSMUST00000227322.2
|
Scel
|
sciellin |
| chr9_-_57342242 | 0.09 |
ENSMUST00000215961.2
|
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chr4_+_151014133 | 0.08 |
ENSMUST00000060901.11
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr7_-_141055044 | 0.08 |
ENSMUST00000043870.9
|
Polr2l
|
polymerase (RNA) II (DNA directed) polypeptide L |
| chr11_-_80030735 | 0.08 |
ENSMUST00000136996.2
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr4_-_143626950 | 0.07 |
ENSMUST00000081645.13
|
Pramel11
|
PRAME like 11 |
| chr11_-_61610772 | 0.07 |
ENSMUST00000151780.8
ENSMUST00000148584.2 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr2_-_90054837 | 0.07 |
ENSMUST00000213994.3
|
Olfr1506
|
olfactory receptor 1506 |
| chr16_+_31918707 | 0.06 |
ENSMUST00000042869.7
|
Cep19
|
centrosomal protein 19 |
| chr13_+_23317325 | 0.05 |
ENSMUST00000227050.2
ENSMUST00000227160.2 ENSMUST00000227741.2 ENSMUST00000226692.2 |
Vmn1r218
|
vomeronasal 1 receptor 218 |
| chr16_-_92156312 | 0.05 |
ENSMUST00000051705.7
|
Kcne1
|
potassium voltage-gated channel, Isk-related subfamily, member 1 |
| chr10_+_62088104 | 0.04 |
ENSMUST00000020278.6
|
Tacr2
|
tachykinin receptor 2 |
| chr14_+_103750814 | 0.04 |
ENSMUST00000095576.5
|
Scel
|
sciellin |
| chr6_+_36364990 | 0.04 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr7_-_126461000 | 0.02 |
ENSMUST00000106343.3
ENSMUST00000205349.2 ENSMUST00000206349.2 |
Ino80e
|
INO80 complex subunit E |
| chr7_+_12034357 | 0.01 |
ENSMUST00000072801.5
ENSMUST00000227672.2 |
Vmn1r82
|
vomeronasal 1 receptor 82 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.9 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.8 | 5.3 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.7 | 6.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.5 | 4.6 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.2 | 7.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 1.1 | 3.4 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 1.1 | 3.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.9 | 7.5 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.9 | 6.9 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.9 | 2.6 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.8 | 3.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.8 | 6.3 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.7 | 2.8 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.7 | 2.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.6 | 3.9 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.6 | 1.9 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.6 | 2.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.6 | 3.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.6 | 1.7 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.6 | 11.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.5 | 1.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 3.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.4 | 3.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.4 | 1.5 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.4 | 8.1 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.4 | 1.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 4.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 2.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 0.9 | GO:0097037 | heme export(GO:0097037) |
| 0.3 | 1.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.3 | 1.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.6 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.3 | 2.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 6.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.3 | 12.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 1.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 4.4 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.2 | 3.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 3.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 0.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 9.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.5 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.2 | 0.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 0.6 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.2 | 0.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 5.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 2.7 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.4 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.1 | 1.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.8 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 1.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 5.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.3 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 1.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.4 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 | 1.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.7 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.8 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 1.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 4.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 2.3 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.0 | 0.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 2.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 4.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 2.6 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 1.5 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 1.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.1 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 1.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 1.3 | 3.9 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 1.1 | 4.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 3.5 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.6 | 3.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 4.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 0.9 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.2 | 3.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 5.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 1.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 5.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 2.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 1.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 0.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 6.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 2.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 9.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 12.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 4.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 25.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.4 | GO:0000793 | condensed chromosome(GO:0000793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.7 | 6.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.6 | 4.8 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.8 | 9.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.8 | 3.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.7 | 5.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.7 | 6.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 3.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.6 | 1.7 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.5 | 2.8 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 3.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 4.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 2.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 1.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 4.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 3.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.9 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 2.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 3.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.5 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 5.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 14.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 5.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.5 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 12.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 9.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 2.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 4.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 2.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 2.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 2.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 11.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 8.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 3.9 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 4.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 15.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 3.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 3.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 6.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 6.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 9.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 1.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 12.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 1.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 5.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 8.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 10.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 5.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.6 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |