Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tbx3
Z-value: 0.66
Transcription factors associated with Tbx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx3
|
ENSMUSG00000018604.19 | Tbx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx3 | mm39_v1_chr5_+_119808722_119808741 | -0.16 | 1.9e-01 | Click! |
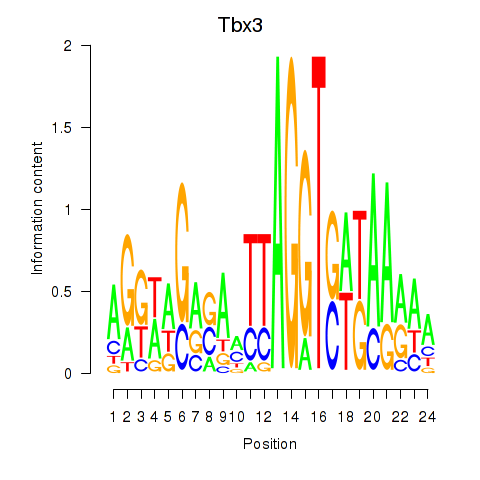
Activity profile of Tbx3 motif
Sorted Z-values of Tbx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_4525426 | 10.43 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr7_-_4525793 | 9.45 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr3_-_122828592 | 6.76 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr2_+_173561208 | 4.75 |
ENSMUST00000073081.6
|
1700010B08Rik
|
RIKEN cDNA 1700010B08 gene |
| chr16_-_13548307 | 4.19 |
ENSMUST00000115807.9
|
Pla2g10
|
phospholipase A2, group X |
| chr16_-_13548833 | 4.16 |
ENSMUST00000023364.7
|
Pla2g10
|
phospholipase A2, group X |
| chr10_+_56253418 | 3.83 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr10_+_127350820 | 3.53 |
ENSMUST00000035735.11
|
Ndufa4l2
|
Ndufa4, mitochondrial complex associated like 2 |
| chr18_-_43526411 | 3.39 |
ENSMUST00000025379.14
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr7_-_43885522 | 2.70 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr12_-_84408576 | 2.42 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr7_+_43885573 | 2.38 |
ENSMUST00000223070.2
ENSMUST00000205530.2 |
Gm36864
|
predicted gene, 36864 |
| chr11_-_100861713 | 2.36 |
ENSMUST00000060792.6
|
Cavin1
|
caveolae associated 1 |
| chr3_-_51184895 | 2.33 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr3_-_87985602 | 2.28 |
ENSMUST00000050258.9
|
Ttc24
|
tetratricopeptide repeat domain 24 |
| chr7_-_43885552 | 2.22 |
ENSMUST00000236952.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chrX_-_133377225 | 2.01 |
ENSMUST00000009740.9
|
Taf7l
|
TATA-box binding protein associated factor 7 like |
| chr5_-_107074110 | 1.93 |
ENSMUST00000117588.8
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chrX_+_126301916 | 1.86 |
ENSMUST00000051530.4
|
4921511C20Rik
|
RIKEN cDNA 4921511C20 gene |
| chr13_-_59853995 | 1.75 |
ENSMUST00000066510.8
ENSMUST00000224469.2 |
Spata31d1a
|
spermatogenesis associated 31 subfamily D, member 1A |
| chr16_+_17437367 | 1.57 |
ENSMUST00000231887.2
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr7_+_28222261 | 1.40 |
ENSMUST00000078364.4
|
Ifnl3
|
interferon lambda 3 |
| chr10_+_80062468 | 1.36 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr19_+_5754395 | 1.16 |
ENSMUST00000052448.4
|
Kcnk7
|
potassium channel, subfamily K, member 7 |
| chr6_-_97037366 | 1.05 |
ENSMUST00000089295.6
|
Tafa4
|
TAFA chemokine like family member 4 |
| chr2_+_154393691 | 0.97 |
ENSMUST00000104928.2
|
Actl10
|
actin-like 10 |
| chr1_-_31261678 | 0.95 |
ENSMUST00000187892.8
ENSMUST00000233331.2 |
4931428L18Rik
|
RIKEN cDNA 4931428L18 gene |
| chr12_+_84408803 | 0.88 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr15_-_97806142 | 0.87 |
ENSMUST00000023119.15
|
Vdr
|
vitamin D (1,25-dihydroxyvitamin D3) receptor |
| chr12_+_84408742 | 0.84 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr8_+_22313550 | 0.37 |
ENSMUST00000078879.7
|
Defb50
|
defensin beta 50 |
| chr2_-_90101376 | 0.27 |
ENSMUST00000216383.2
|
Olfr1271
|
olfactory receptor 1271 |
| chr11_+_73241609 | 0.23 |
ENSMUST00000120137.3
|
Olfr20
|
olfactory receptor 20 |
| chr10_-_18619439 | 0.23 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr19_+_7471398 | 0.18 |
ENSMUST00000170373.9
ENSMUST00000236308.2 ENSMUST00000235557.2 |
Atl3
|
atlastin GTPase 3 |
| chr14_+_67982630 | 0.18 |
ENSMUST00000223929.2
ENSMUST00000111095.4 |
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr19_-_10434837 | 0.13 |
ENSMUST00000171400.4
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr3_+_60380243 | 0.11 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr16_+_17437205 | 0.06 |
ENSMUST00000006053.13
ENSMUST00000231311.2 ENSMUST00000163476.10 ENSMUST00000231257.2 ENSMUST00000165363.10 ENSMUST00000232043.2 ENSMUST00000090159.13 ENSMUST00000232442.2 |
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr16_-_19241884 | 0.04 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr10_-_18619658 | 0.02 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr1_+_39574829 | 0.02 |
ENSMUST00000003219.14
|
Cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr7_-_119744509 | 0.02 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 19.9 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 2.8 | 8.3 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 1.3 | 3.8 | GO:0010232 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.6 | 2.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 0.9 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.2 | 1.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 3.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 6.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 2.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 19.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 3.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 3.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 2.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 3.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 5.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 2.4 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 19.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.4 | 6.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 3.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 3.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 0.9 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.2 | 8.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 1.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 3.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 2.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 19.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |