Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
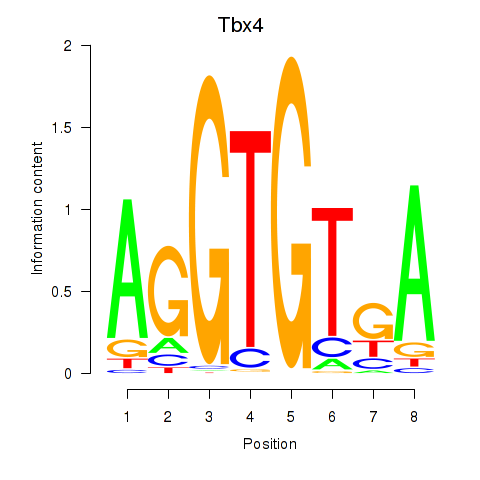
Results for Tbx4
Z-value: 0.74
Transcription factors associated with Tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx4
|
ENSMUSG00000000094.13 | Tbx4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx4 | mm39_v1_chr11_+_85780807_85780889 | 0.64 | 1.2e-09 | Click! |
Activity profile of Tbx4 motif
Sorted Z-values of Tbx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_72281587 | 6.59 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr9_-_99592116 | 5.77 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr9_-_99592058 | 5.68 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr2_-_147888816 | 5.53 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr2_+_164404499 | 5.41 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr19_+_20579322 | 4.98 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr7_+_79460475 | 4.50 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr2_-_147887810 | 4.00 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr7_-_103463120 | 3.92 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr7_-_79443536 | 3.60 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr7_-_119694400 | 2.81 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr15_+_38219447 | 2.80 |
ENSMUST00000081966.5
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr5_+_17779721 | 2.79 |
ENSMUST00000169603.2
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr14_-_70449438 | 2.79 |
ENSMUST00000227929.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr4_-_133746138 | 2.74 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr4_+_148085179 | 2.48 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr1_+_40619215 | 2.46 |
ENSMUST00000027233.9
|
Slc9a4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4 |
| chr10_+_86541675 | 2.35 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr15_-_11905697 | 2.27 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr4_-_19922599 | 2.21 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr4_+_43641262 | 2.01 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr11_-_94398162 | 1.90 |
ENSMUST00000040692.9
|
Mycbpap
|
MYCBP associated protein |
| chr7_-_81216687 | 1.86 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr16_+_62635039 | 1.85 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr8_+_55024446 | 1.78 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr19_-_28941264 | 1.77 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr2_+_103254465 | 1.73 |
ENSMUST00000171693.8
|
Elf5
|
E74-like factor 5 |
| chr3_-_30194559 | 1.70 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr6_+_139987275 | 1.70 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr4_-_46404224 | 1.64 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr6_+_15196950 | 1.57 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr18_+_32948436 | 1.54 |
ENSMUST00000025237.5
|
Tslp
|
thymic stromal lymphopoietin |
| chr15_-_78687216 | 1.51 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr2_+_103254401 | 1.51 |
ENSMUST00000028609.14
|
Elf5
|
E74-like factor 5 |
| chr18_-_65527078 | 1.46 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr18_-_60724855 | 1.46 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr7_-_100307601 | 1.41 |
ENSMUST00000138830.2
ENSMUST00000107044.10 ENSMUST00000116287.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr19_-_45224251 | 1.40 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr19_-_37184692 | 1.39 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr10_+_97528915 | 1.37 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr7_+_130179063 | 1.35 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_+_119356318 | 1.33 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr6_-_139987135 | 1.31 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr8_+_46338557 | 1.30 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr11_-_23720953 | 1.29 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr7_-_73663422 | 1.27 |
ENSMUST00000026896.10
ENSMUST00000191970.2 |
St8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr4_+_117706559 | 1.23 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr10_+_87697155 | 1.19 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr16_-_32617783 | 1.16 |
ENSMUST00000115116.8
ENSMUST00000041123.9 |
Muc20
|
mucin 20 |
| chr1_+_74582044 | 1.15 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr9_+_102885156 | 1.14 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr16_+_87350202 | 1.13 |
ENSMUST00000026700.8
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr7_-_100307571 | 1.10 |
ENSMUST00000107043.8
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_-_14420725 | 1.09 |
ENSMUST00000041168.6
ENSMUST00000238680.2 |
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr4_+_117706390 | 1.08 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_32498997 | 1.07 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chrX_+_167819606 | 1.05 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr14_-_30665232 | 1.05 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr19_+_37686240 | 1.04 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr1_+_59563453 | 0.97 |
ENSMUST00000186434.3
|
Gm973
|
predicted gene 973 |
| chr1_-_126758520 | 0.96 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr11_-_22236795 | 0.95 |
ENSMUST00000180360.8
ENSMUST00000109563.9 |
Ehbp1
|
EH domain binding protein 1 |
| chr2_+_103800553 | 0.94 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr6_+_42263609 | 0.93 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr1_-_126758369 | 0.91 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr7_+_4240697 | 0.89 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr9_-_107486381 | 0.89 |
ENSMUST00000102531.7
ENSMUST00000102530.8 ENSMUST00000195057.2 ENSMUST00000102532.10 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr8_+_46338498 | 0.88 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr4_-_72770859 | 0.87 |
ENSMUST00000179234.2
ENSMUST00000078617.5 |
Aldoart1
|
aldolase 1 A, retrogene 1 |
| chr13_+_81034214 | 0.82 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr3_+_40905066 | 0.79 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr9_+_71123061 | 0.76 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chrX_+_132809166 | 0.75 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_-_108529375 | 0.75 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr12_-_31684588 | 0.74 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr17_+_35878332 | 0.74 |
ENSMUST00000044326.5
|
2300002M23Rik
|
RIKEN cDNA 2300002M23 gene |
| chr13_-_89890609 | 0.72 |
ENSMUST00000109546.9
|
Vcan
|
versican |
| chr9_+_69902697 | 0.71 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr6_-_52141796 | 0.71 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr19_-_10282218 | 0.67 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr3_+_40905216 | 0.65 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr11_+_95304903 | 0.63 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr13_-_96678844 | 0.62 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr15_+_10952418 | 0.60 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr18_+_65715460 | 0.57 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr2_+_138120401 | 0.57 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr6_-_126143710 | 0.56 |
ENSMUST00000050484.9
|
Ntf3
|
neurotrophin 3 |
| chr6_+_42263644 | 0.53 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr17_+_35354655 | 0.52 |
ENSMUST00000174478.8
ENSMUST00000174281.9 ENSMUST00000173550.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr11_+_96214078 | 0.49 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr9_-_40915858 | 0.48 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr12_-_56392646 | 0.48 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr11_-_83959175 | 0.47 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr10_-_86541349 | 0.46 |
ENSMUST00000020238.14
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr4_-_42665763 | 0.46 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr13_+_113346193 | 0.46 |
ENSMUST00000038144.9
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr4_-_42168603 | 0.45 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_+_42950367 | 0.45 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr3_-_75464066 | 0.45 |
ENSMUST00000162138.2
ENSMUST00000029424.12 ENSMUST00000161137.8 |
Pdcd10
|
programmed cell death 10 |
| chr16_-_32630847 | 0.44 |
ENSMUST00000179384.3
|
Smbd1
|
somatomedin B domain containing 1 |
| chrM_+_9459 | 0.44 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr7_+_89779564 | 0.41 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_78068931 | 0.41 |
ENSMUST00000147819.8
|
Tlcd1
|
TLC domain containing 1 |
| chr16_+_84631789 | 0.41 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr11_+_23256883 | 0.39 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr18_+_5591864 | 0.39 |
ENSMUST00000025081.13
ENSMUST00000159390.8 |
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chrM_+_10167 | 0.38 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_+_136733421 | 0.37 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr6_+_29398919 | 0.36 |
ENSMUST00000181464.8
ENSMUST00000180829.8 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr14_+_45588632 | 0.36 |
ENSMUST00000228818.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr9_-_102231884 | 0.36 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chrX_+_158623460 | 0.34 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_-_99494134 | 0.33 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr9_+_108174052 | 0.33 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr16_-_50151350 | 0.32 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chrM_+_9870 | 0.32 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr9_-_40915895 | 0.31 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr3_-_121056944 | 0.30 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr7_-_28001624 | 0.30 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr7_-_141850806 | 0.30 |
ENSMUST00000084413.7
ENSMUST00000188274.2 |
Krtap5-1
|
keratin associated protein 5-1 |
| chr16_+_84631956 | 0.29 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr13_-_96678987 | 0.29 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr3_+_51323383 | 0.27 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr18_-_34505544 | 0.26 |
ENSMUST00000236887.2
|
Reep5
|
receptor accessory protein 5 |
| chr9_+_107468146 | 0.26 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr7_-_28078671 | 0.25 |
ENSMUST00000209061.2
|
Zfp36
|
zinc finger protein 36 |
| chr5_+_120651158 | 0.24 |
ENSMUST00000111889.2
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr6_-_145811274 | 0.24 |
ENSMUST00000032386.11
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr7_-_115423934 | 0.22 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_+_52747855 | 0.22 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr7_+_46445512 | 0.21 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr7_+_45890379 | 0.21 |
ENSMUST00000164538.2
|
Otog
|
otogelin |
| chr8_-_46605196 | 0.20 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr11_+_121128042 | 0.19 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor |
| chr1_+_171156568 | 0.19 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr14_-_55762395 | 0.19 |
ENSMUST00000228287.2
|
Nrl
|
neural retina leucine zipper gene |
| chr12_-_11258973 | 0.18 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr18_+_36414122 | 0.18 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr9_+_106247943 | 0.16 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr8_-_32408786 | 0.15 |
ENSMUST00000239000.2
|
Nrg1
|
neuregulin 1 |
| chr11_-_106192627 | 0.13 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr14_-_55762432 | 0.13 |
ENSMUST00000062232.15
|
Nrl
|
neural retina leucine zipper gene |
| chr14_+_45588509 | 0.11 |
ENSMUST00000226873.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr16_-_23807602 | 0.11 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr11_-_117859997 | 0.10 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr2_+_54326329 | 0.10 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr6_-_145811028 | 0.10 |
ENSMUST00000111703.2
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr14_-_55762416 | 0.09 |
ENSMUST00000178694.3
|
Nrl
|
neural retina leucine zipper gene |
| chr13_-_111626562 | 0.09 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr17_-_49871291 | 0.08 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr8_-_32408380 | 0.07 |
ENSMUST00000208497.3
ENSMUST00000207584.3 |
Nrg1
|
neuregulin 1 |
| chr11_-_99511257 | 0.07 |
ENSMUST00000073853.3
|
Gm11562
|
predicted gene 11562 |
| chr14_+_53574579 | 0.07 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr13_-_43457626 | 0.04 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr2_+_130038463 | 0.04 |
ENSMUST00000166774.3
|
Tmc2
|
transmembrane channel-like gene family 2 |
| chrX_+_41239872 | 0.04 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr11_-_98040377 | 0.03 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr8_-_32408864 | 0.03 |
ENSMUST00000073884.7
ENSMUST00000238812.2 |
Nrg1
|
neuregulin 1 |
| chr11_-_69791774 | 0.03 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr9_+_72952115 | 0.02 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr4_-_133399909 | 0.02 |
ENSMUST00000062118.11
ENSMUST00000067902.13 |
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr13_+_96679233 | 0.01 |
ENSMUST00000077672.12
ENSMUST00000109444.3 |
Cert1
|
ceramide transporter 1 |
| chr14_+_52131067 | 0.01 |
ENSMUST00000047726.12
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr19_+_55882942 | 0.00 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.6 | 11.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.2 | 3.6 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.8 | 2.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.6 | 5.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.6 | 2.5 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.6 | 4.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.5 | 3.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.5 | 1.4 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 2.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 2.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 0.8 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 2.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 2.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 2.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 2.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.7 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.2 | 1.2 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 0.7 | GO:0099541 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.2 | 2.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.5 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 0.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 1.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 1.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.8 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 1.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 1.9 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.1 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.2 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.7 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 2.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 1.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.1 | 5.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.6 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.5 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.1 | 0.9 | GO:0097067 | negative regulation of erythrocyte differentiation(GO:0045647) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 1.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 1.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:2001015 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 1.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.2 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 6.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 2.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 2.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 2.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 2.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 11.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.0 | 5.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.0 | 3.9 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.7 | 4.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 1.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 2.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.8 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 3.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 1.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 2.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 10.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 1.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 2.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.2 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 1.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 3.6 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 11.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 4.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 3.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 7.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 9.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 11.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 9.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |