Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tbx5
Z-value: 0.95
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSMUSG00000018263.12 | Tbx5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx5 | mm39_v1_chr5_+_119970733_119970780 | -0.04 | 7.5e-01 | Click! |
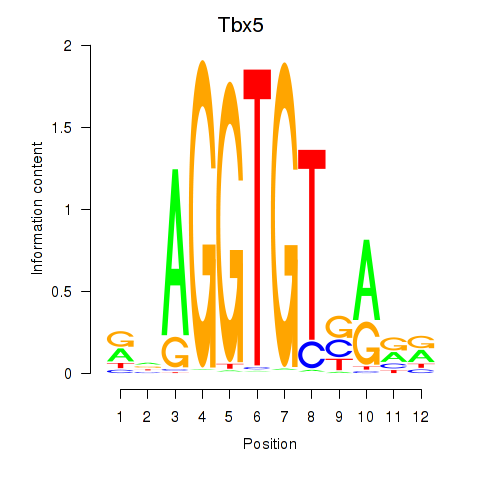
Activity profile of Tbx5 motif
Sorted Z-values of Tbx5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_100396635 | 5.15 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr16_-_10614679 | 4.39 |
ENSMUST00000023144.6
|
Prm1
|
protamine 1 |
| chr7_-_44888532 | 4.02 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr4_-_133746138 | 4.02 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chrX_+_99773523 | 3.97 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr3_-_92493507 | 3.94 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chrX_+_99773784 | 3.80 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr10_+_97528915 | 3.54 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr19_-_6902735 | 3.49 |
ENSMUST00000057716.6
|
Catsperz
|
cation channel sperm associated auxiliary subunit zeta |
| chr7_-_44888220 | 3.40 |
ENSMUST00000210372.2
ENSMUST00000209779.2 ENSMUST00000098461.10 ENSMUST00000211373.2 |
Cd37
|
CD37 antigen |
| chr6_+_125026915 | 3.36 |
ENSMUST00000088294.12
ENSMUST00000032481.8 |
Acrbp
|
proacrosin binding protein |
| chr2_-_32277773 | 3.34 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr19_-_6902698 | 3.33 |
ENSMUST00000237380.2
|
Catsperz
|
cation channel sperm associated auxiliary subunit zeta |
| chr5_-_100867520 | 3.32 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr15_-_78280099 | 3.30 |
ENSMUST00000229878.2
ENSMUST00000165170.8 ENSMUST00000074380.14 |
Tex33
|
testis expressed 33 |
| chr7_+_28682253 | 3.24 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr15_-_103163860 | 3.20 |
ENSMUST00000075192.13
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr6_-_136638926 | 3.11 |
ENSMUST00000032336.7
|
Plbd1
|
phospholipase B domain containing 1 |
| chr1_-_156936197 | 3.05 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr6_-_135287372 | 3.02 |
ENSMUST00000050471.4
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr11_-_83412952 | 2.94 |
ENSMUST00000019069.4
|
Heatr9
|
HEAT repeat containing 9 |
| chr15_-_89294434 | 2.86 |
ENSMUST00000109314.9
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr8_-_105019806 | 2.81 |
ENSMUST00000212492.2
ENSMUST00000034344.10 |
Cmtm2a
|
CKLF-like MARVEL transmembrane domain containing 2A |
| chr11_-_118233326 | 2.79 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr3_-_95214102 | 2.76 |
ENSMUST00000107183.8
ENSMUST00000164406.8 ENSMUST00000123365.2 |
Anxa9
|
annexin A9 |
| chr17_+_48571298 | 2.72 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr6_+_41661356 | 2.69 |
ENSMUST00000031900.6
|
Llcfc1
|
LLLL and CFNLAS motif containing 1 |
| chr18_-_46345661 | 2.65 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr8_+_84116507 | 2.61 |
ENSMUST00000109831.3
|
Clgn
|
calmegin |
| chr4_-_41517326 | 2.59 |
ENSMUST00000030152.13
ENSMUST00000095126.5 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr2_-_32278245 | 2.57 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr7_-_44888465 | 2.56 |
ENSMUST00000210078.2
|
Cd37
|
CD37 antigen |
| chr14_-_56322654 | 2.51 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr3_-_95214443 | 2.51 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr2_+_103800459 | 2.49 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr2_+_103800553 | 2.49 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr2_+_24076481 | 2.46 |
ENSMUST00000057567.3
|
Il1f9
|
interleukin 1 family, member 9 |
| chr8_+_84116463 | 2.40 |
ENSMUST00000002259.13
|
Clgn
|
calmegin |
| chr6_-_52168675 | 2.39 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr8_-_106660470 | 2.36 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr1_-_131172903 | 2.30 |
ENSMUST00000027688.15
ENSMUST00000112442.2 |
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr3_-_20329823 | 2.23 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chrX_-_36253309 | 2.19 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr12_+_51640097 | 2.19 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr6_+_139987275 | 2.17 |
ENSMUST00000043797.6
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr18_-_55123153 | 2.14 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr1_-_172156884 | 2.12 |
ENSMUST00000062387.8
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr3_-_92514799 | 2.09 |
ENSMUST00000195278.2
|
2210017I01Rik
|
RIKEN cDNA 2210017I01 gene |
| chr15_-_82917495 | 2.08 |
ENSMUST00000231165.2
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chr6_-_73446560 | 2.06 |
ENSMUST00000070163.6
|
4931417E11Rik
|
RIKEN cDNA 4931417E11 gene |
| chr19_-_46315543 | 2.04 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr6_+_125026865 | 2.02 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr11_+_70529944 | 2.00 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr6_-_49191891 | 1.98 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr14_-_35533922 | 1.90 |
ENSMUST00000043266.7
|
4930596D02Rik
|
RIKEN cDNA 4930596D02 gene |
| chr6_-_139987135 | 1.86 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1 |
| chr15_+_73620213 | 1.79 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr11_-_69791712 | 1.76 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_-_69791756 | 1.72 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr8_+_34007333 | 1.71 |
ENSMUST00000124496.8
|
Tex15
|
testis expressed gene 15 |
| chr19_-_28941264 | 1.66 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr6_+_70549568 | 1.65 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr10_+_60182630 | 1.65 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr14_-_55950545 | 1.63 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr14_-_51295099 | 1.63 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr3_+_127583450 | 1.63 |
ENSMUST00000054483.14
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr7_+_44145987 | 1.62 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr15_-_89310060 | 1.62 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr11_+_6339442 | 1.60 |
ENSMUST00000109786.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr9_+_95441652 | 1.59 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr6_-_122463422 | 1.58 |
ENSMUST00000068242.9
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr12_-_31684588 | 1.58 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr9_-_106076389 | 1.57 |
ENSMUST00000140761.9
|
Ppm1m
|
protein phosphatase 1M |
| chr4_+_9844349 | 1.56 |
ENSMUST00000057613.3
|
Gdf6
|
growth differentiation factor 6 |
| chr1_-_172156828 | 1.56 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr15_-_9748863 | 1.55 |
ENSMUST00000159368.2
ENSMUST00000159093.8 ENSMUST00000162780.8 ENSMUST00000160236.8 ENSMUST00000208854.2 ENSMUST00000041840.14 |
Spef2
|
sperm flagellar 2 |
| chr3_-_92758591 | 1.54 |
ENSMUST00000054426.5
|
Lce1l
|
late cornified envelope 1L |
| chr1_+_134313676 | 1.54 |
ENSMUST00000162187.2
|
Mgat4f
|
MGAT4 family, member F |
| chr1_+_132119169 | 1.53 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr6_-_122463316 | 1.51 |
ENSMUST00000205114.2
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr5_-_77262968 | 1.51 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr4_+_114916703 | 1.47 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr3_+_75464837 | 1.47 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr8_-_46605196 | 1.45 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr11_-_69791774 | 1.45 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chrX_+_158623460 | 1.44 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_+_6339330 | 1.44 |
ENSMUST00000012612.11
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr2_-_181333597 | 1.43 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr4_+_33031433 | 1.43 |
ENSMUST00000029944.13
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr1_-_71454041 | 1.43 |
ENSMUST00000087268.7
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr11_-_3813895 | 1.43 |
ENSMUST00000070552.14
|
Osbp2
|
oxysterol binding protein 2 |
| chr4_+_154954042 | 1.40 |
ENSMUST00000079269.14
ENSMUST00000163732.8 ENSMUST00000080559.13 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr3_-_92922976 | 1.40 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr11_-_106192627 | 1.40 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr6_-_40562700 | 1.39 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr7_+_81823546 | 1.39 |
ENSMUST00000179318.8
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr3_+_90173813 | 1.39 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr14_-_70855980 | 1.38 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr11_-_87766350 | 1.38 |
ENSMUST00000049768.4
|
Epx
|
eosinophil peroxidase |
| chr1_-_75195889 | 1.37 |
ENSMUST00000186213.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr7_+_43086432 | 1.37 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr7_-_28001624 | 1.36 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr15_-_89309998 | 1.36 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr3_-_75464066 | 1.35 |
ENSMUST00000162138.2
ENSMUST00000029424.12 ENSMUST00000161137.8 |
Pdcd10
|
programmed cell death 10 |
| chr3_+_127583550 | 1.33 |
ENSMUST00000163775.6
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr2_-_38602207 | 1.33 |
ENSMUST00000112883.8
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr19_-_41373526 | 1.32 |
ENSMUST00000059672.9
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr11_+_6339061 | 1.32 |
ENSMUST00000109787.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr12_-_114057841 | 1.32 |
ENSMUST00000103471.2
ENSMUST00000195884.2 |
Ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr7_+_43086554 | 1.31 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr8_+_84874654 | 1.28 |
ENSMUST00000143833.8
ENSMUST00000118856.8 |
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr2_-_68302612 | 1.27 |
ENSMUST00000102715.4
|
Stk39
|
serine/threonine kinase 39 |
| chr1_-_132318039 | 1.26 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr9_-_62888156 | 1.24 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr10_+_56253418 | 1.23 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr17_+_71859026 | 1.23 |
ENSMUST00000124001.8
ENSMUST00000167641.8 ENSMUST00000064420.12 |
Spdya
|
speedy/RINGO cell cycle regulator family, member A |
| chr7_+_44146029 | 1.23 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr8_+_23525101 | 1.19 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr1_+_181404124 | 1.18 |
ENSMUST00000208001.2
|
Dnah14
|
dynein, axonemal, heavy chain 14 |
| chr9_-_107109108 | 1.17 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr10_+_86541675 | 1.17 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr18_+_82932747 | 1.14 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr15_-_99864936 | 1.14 |
ENSMUST00000100209.6
|
Fam186a
|
family with sequence similarity 186, member A |
| chr12_-_113700190 | 1.13 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr7_-_25488060 | 1.13 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr15_+_100659729 | 1.12 |
ENSMUST00000161564.2
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr6_+_30639217 | 1.12 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr2_+_28403255 | 1.11 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr8_+_84874881 | 1.08 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr11_-_23720953 | 1.08 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr11_+_113575208 | 1.08 |
ENSMUST00000042227.15
ENSMUST00000123466.2 ENSMUST00000106621.4 |
D11Wsu47e
|
DNA segment, Chr 11, Wayne State University 47, expressed |
| chr1_-_92830604 | 1.08 |
ENSMUST00000160548.8
ENSMUST00000112998.2 |
Ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr15_+_100659622 | 1.07 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr6_+_48695535 | 1.06 |
ENSMUST00000127537.6
ENSMUST00000052503.8 |
Gm28053
Gimap7
|
predicted gene, 28053 GTPase, IMAP family member 7 |
| chr7_+_24990596 | 1.05 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr1_+_63312420 | 1.04 |
ENSMUST00000239483.2
ENSMUST00000114132.8 ENSMUST00000126932.2 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr14_-_70414236 | 1.02 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr7_-_101749433 | 1.02 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr12_-_113823290 | 1.01 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr2_-_91013193 | 1.01 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chr7_+_81508741 | 1.00 |
ENSMUST00000041890.8
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr19_+_23881821 | 1.00 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr5_-_148336711 | 0.98 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_-_144174066 | 0.98 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr6_-_84564623 | 0.97 |
ENSMUST00000205228.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr13_-_113317431 | 0.97 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chr11_+_54194831 | 0.97 |
ENSMUST00000000145.12
ENSMUST00000138515.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr4_+_132291369 | 0.96 |
ENSMUST00000070690.8
|
Ptafr
|
platelet-activating factor receptor |
| chr5_+_98328723 | 0.96 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chr8_+_34006758 | 0.96 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr11_+_115044966 | 0.96 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr12_+_69954506 | 0.96 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr6_-_84564972 | 0.96 |
ENSMUST00000204109.2
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr4_+_33031342 | 0.96 |
ENSMUST00000124992.8
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr6_+_35229628 | 0.95 |
ENSMUST00000130875.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr7_-_109037992 | 0.95 |
ENSMUST00000121748.8
|
Stk33
|
serine/threonine kinase 33 |
| chrX_+_73220858 | 0.95 |
ENSMUST00000010127.12
|
Tktl1
|
transketolase-like 1 |
| chr11_+_54195006 | 0.94 |
ENSMUST00000108904.10
ENSMUST00000108905.10 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr1_-_161616031 | 0.94 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr1_+_9868332 | 0.93 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr10_+_84412490 | 0.93 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr15_+_103411689 | 0.92 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr17_-_7620095 | 0.92 |
ENSMUST00000115747.3
|
Ttll2
|
tubulin tyrosine ligase-like family, member 2 |
| chr9_-_20978389 | 0.92 |
ENSMUST00000115494.3
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr4_+_83443777 | 0.91 |
ENSMUST00000053414.13
ENSMUST00000231339.2 ENSMUST00000126429.8 |
Ccdc171
|
coiled-coil domain containing 171 |
| chr7_-_109038282 | 0.91 |
ENSMUST00000106745.9
ENSMUST00000090414.11 ENSMUST00000141210.2 |
Stk33
|
serine/threonine kinase 33 |
| chr3_+_92315290 | 0.90 |
ENSMUST00000047264.3
|
Sprr2i
|
small proline-rich protein 2I |
| chr3_-_37180093 | 0.90 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr7_-_27095964 | 0.89 |
ENSMUST00000108363.8
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chrX_+_99669343 | 0.88 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr4_-_43045685 | 0.88 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr15_-_75760602 | 0.87 |
ENSMUST00000184858.2
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr7_-_109038202 | 0.86 |
ENSMUST00000121378.2
|
Stk33
|
serine/threonine kinase 33 |
| chrX_-_59498719 | 0.85 |
ENSMUST00000114687.9
|
Gm7073
|
predicted gene 7073 |
| chr4_-_133329479 | 0.85 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr15_-_100583044 | 0.84 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr17_+_47815968 | 0.84 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr7_+_89779564 | 0.83 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_-_19595221 | 0.83 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr17_-_32255287 | 0.82 |
ENSMUST00000238192.2
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr13_+_13764982 | 0.82 |
ENSMUST00000110559.3
|
Lyst
|
lysosomal trafficking regulator |
| chrX_+_41578568 | 0.82 |
ENSMUST00000197237.2
|
Tex13c2
|
TEX13 family member C2 |
| chr5_-_142892457 | 0.81 |
ENSMUST00000167721.8
ENSMUST00000163829.2 ENSMUST00000100497.11 |
Actb
|
actin, beta |
| chr1_-_74323795 | 0.81 |
ENSMUST00000178235.8
ENSMUST00000006462.14 |
Aamp
|
angio-associated migratory protein |
| chr7_-_112968533 | 0.81 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr11_-_82761954 | 0.81 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr10_+_80538115 | 0.80 |
ENSMUST00000218184.2
|
Izumo4
|
IZUMO family member 4 |
| chr12_-_113733922 | 0.80 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr19_-_37184692 | 0.79 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr11_-_46203047 | 0.79 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_74640706 | 0.79 |
ENSMUST00000087186.11
|
Stk36
|
serine/threonine kinase 36 |
| chr17_+_47816074 | 0.79 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr1_+_132243849 | 0.79 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr1_+_74640590 | 0.78 |
ENSMUST00000087183.11
ENSMUST00000148456.8 ENSMUST00000113694.8 |
Stk36
|
serine/threonine kinase 36 |
| chr7_+_44146012 | 0.78 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr17_+_47816042 | 0.77 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr7_+_19197192 | 0.77 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr7_-_24771717 | 0.76 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr10_+_69048506 | 0.76 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_-_167112784 | 0.75 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.1 | 3.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 1.0 | 1.0 | GO:1902941 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.9 | 10.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.7 | 4.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.6 | 2.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.5 | 9.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 1.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 3.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 1.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.5 | 1.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.5 | 2.8 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.5 | 1.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.4 | 1.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.4 | 1.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.4 | 2.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 1.2 | GO:0010232 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.4 | 1.6 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.4 | 1.9 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.3 | 2.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 1.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 1.9 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.3 | 5.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 12.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 0.8 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 0.8 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.3 | 1.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 0.8 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.3 | 0.8 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 0.7 | GO:0032262 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.2 | 1.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 1.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 2.5 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 1.1 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.2 | 1.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 2.4 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 2.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.2 | 5.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 0.7 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.2 | 0.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 0.5 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.8 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 3.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 1.9 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 1.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 4.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 1.8 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.3 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 5.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 1.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.2 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.9 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.4 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 2.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.4 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.9 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 1.4 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.1 | 0.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.5 | GO:0046642 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 1.3 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 1.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 6.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.7 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 1.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 1.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.7 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.0 | 1.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 2.4 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 1.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 3.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 1.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 2.2 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 1.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.4 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.4 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 2.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 2.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 1.5 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.5 | 6.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 4.1 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 4.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 1.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 2.9 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 2.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 9.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 4.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 4.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 1.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 6.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 1.1 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 1.0 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 3.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 2.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 4.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.8 | 3.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.5 | 1.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.4 | 3.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 3.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.3 | 1.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 0.9 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 4.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 0.8 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 3.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 5.0 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.2 | 1.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 1.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.4 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.1 | 0.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 5.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 3.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 2.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 2.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.9 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 5.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 2.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 3.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 3.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 6.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 2.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 2.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 4.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 3.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 2.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 5.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |