Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
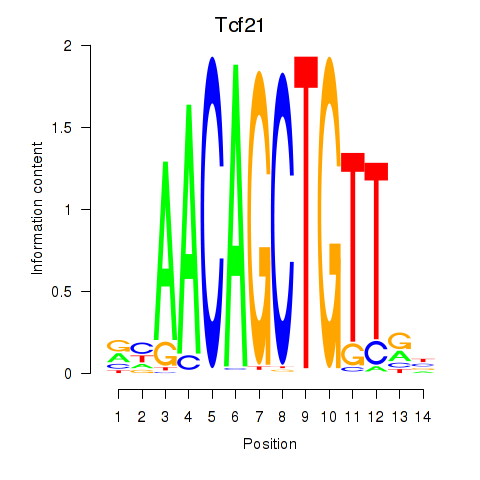
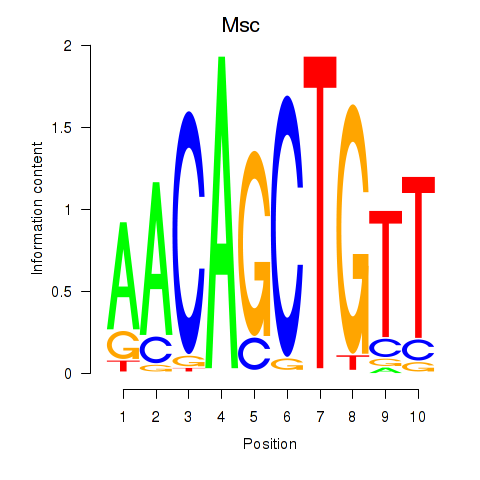
Results for Tcf21_Msc
Z-value: 1.46
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf21
|
ENSMUSG00000045680.9 | Tcf21 |
|
Msc
|
ENSMUSG00000025930.7 | Msc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msc | mm39_v1_chr1_-_14826208_14826222 | 0.40 | 4.2e-04 | Click! |
| Tcf21 | mm39_v1_chr10_-_22696025_22696073 | 0.16 | 1.7e-01 | Click! |
Activity profile of Tcf21_Msc motif
Sorted Z-values of Tcf21_Msc motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf21_Msc
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_92195873 | 14.71 |
ENSMUST00000090872.7
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr7_+_27770655 | 12.74 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr6_-_3494587 | 11.83 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr13_-_100922910 | 10.92 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr14_-_31299275 | 10.84 |
ENSMUST00000112027.9
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr2_-_169973076 | 10.46 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr6_-_70237939 | 9.76 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr14_-_56809287 | 9.61 |
ENSMUST00000065302.8
|
Cenpj
|
centromere protein J |
| chr6_+_68402550 | 9.36 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr7_-_127593003 | 8.55 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr3_+_96465265 | 8.42 |
ENSMUST00000074519.13
ENSMUST00000049093.8 |
Txnip
|
thioredoxin interacting protein |
| chr6_+_70192384 | 7.83 |
ENSMUST00000103383.3
|
Igkv6-25
|
immunoglobulin kappa chain variable 6-25 |
| chr8_+_21624626 | 7.37 |
ENSMUST00000098898.5
|
Defa30
|
defensin, alpha, 30 |
| chr11_+_53661251 | 7.23 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr4_-_149539428 | 7.01 |
ENSMUST00000030848.3
|
Rbp7
|
retinol binding protein 7, cellular |
| chr4_+_11156411 | 6.97 |
ENSMUST00000029865.4
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1 |
| chr4_-_132990362 | 6.91 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr19_+_11493825 | 6.77 |
ENSMUST00000163078.8
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chrX_+_100492684 | 6.73 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr9_-_114610879 | 6.73 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_-_190950129 | 6.67 |
ENSMUST00000195117.2
|
Atf3
|
activating transcription factor 3 |
| chr2_-_173060647 | 6.42 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr11_-_99045894 | 6.40 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr1_-_192880260 | 6.32 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr11_-_100036792 | 6.26 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr5_+_123214332 | 6.18 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr3_+_95496239 | 6.03 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr13_-_74210360 | 5.97 |
ENSMUST00000222609.2
ENSMUST00000036456.8 |
Cep72
|
centrosomal protein 72 |
| chr4_-_87724533 | 5.96 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr19_-_11313471 | 5.95 |
ENSMUST00000056035.9
ENSMUST00000067532.11 |
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr6_-_70383976 | 5.91 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr8_-_123159663 | 5.73 |
ENSMUST00000017604.10
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr7_+_142558783 | 5.68 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr14_+_28740162 | 5.68 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr11_-_40624200 | 5.67 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr3_+_95496270 | 5.65 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr16_+_57173456 | 5.65 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_-_70412460 | 5.62 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr7_+_142025575 | 5.59 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr8_-_123159639 | 5.59 |
ENSMUST00000212600.2
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr6_+_112436466 | 5.55 |
ENSMUST00000075477.8
|
Cav3
|
caveolin 3 |
| chr7_-_81216687 | 5.53 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr6_+_145091196 | 5.46 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr18_+_89215575 | 5.44 |
ENSMUST00000097496.4
ENSMUST00000236452.2 |
Cd226
|
CD226 antigen |
| chr17_-_43813664 | 5.31 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr7_+_142025817 | 5.27 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr7_+_142558837 | 5.21 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr18_+_4994600 | 5.07 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr15_+_78810919 | 4.94 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr1_-_69726384 | 4.87 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr5_-_98178834 | 4.82 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2 |
| chr7_-_140697719 | 4.79 |
ENSMUST00000067836.9
|
Ano9
|
anoctamin 9 |
| chr10_+_79690452 | 4.77 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr4_-_87724512 | 4.72 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_-_70435020 | 4.64 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr15_-_73579236 | 4.59 |
ENSMUST00000064166.5
|
Gpr20
|
G protein-coupled receptor 20 |
| chr4_-_137157824 | 4.52 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chrX_-_105528972 | 4.45 |
ENSMUST00000140707.2
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr12_+_24758240 | 4.41 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr15_-_101912917 | 4.40 |
ENSMUST00000023952.10
|
Krt8
|
keratin 8 |
| chr3_+_106393348 | 4.25 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr7_+_79675727 | 4.24 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr6_-_70051586 | 4.22 |
ENSMUST00000103377.3
|
Igkv6-32
|
immunoglobulin kappa variable 6-32 |
| chr7_-_126303947 | 4.14 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr3_-_100396635 | 4.00 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr11_+_70548622 | 3.85 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr1_+_130793406 | 3.84 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr16_+_96001650 | 3.83 |
ENSMUST00000048770.16
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr11_+_61017573 | 3.74 |
ENSMUST00000010286.8
ENSMUST00000146033.8 ENSMUST00000139422.8 |
Tnfrsf13b
|
tumor necrosis factor receptor superfamily, member 13b |
| chr7_-_4517367 | 3.74 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr1_-_133352115 | 3.74 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr10_+_79690492 | 3.71 |
ENSMUST00000171599.8
ENSMUST00000095457.11 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr8_-_25592001 | 3.69 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr19_+_41017714 | 3.59 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr17_-_48739874 | 3.57 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr2_-_35322581 | 3.55 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr16_-_4377640 | 3.53 |
ENSMUST00000005862.9
|
Tfap4
|
transcription factor AP4 |
| chr3_+_87813624 | 3.46 |
ENSMUST00000005017.15
|
Hdgf
|
heparin binding growth factor |
| chr5_+_103902426 | 3.44 |
ENSMUST00000153165.8
ENSMUST00000031256.6 |
Aff1
|
AF4/FMR2 family, member 1 |
| chr6_-_69678271 | 3.34 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr5_+_64317550 | 3.32 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr14_-_20231871 | 3.32 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr7_-_16549394 | 3.29 |
ENSMUST00000206259.2
|
Fkrp
|
fukutin related protein |
| chr9_+_77543776 | 3.27 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr5_+_31274046 | 3.26 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr15_+_80507671 | 3.24 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr7_-_126046814 | 3.23 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr8_+_46338498 | 3.20 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr11_-_5753693 | 3.19 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr1_-_36574801 | 3.19 |
ENSMUST00000054665.10
ENSMUST00000194853.3 |
Ankrd23
|
ankyrin repeat domain 23 |
| chr1_-_36578439 | 3.17 |
ENSMUST00000191849.6
|
Gm42417
|
predicted gene, 42417 |
| chr3_+_40755211 | 3.17 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chrX_-_100310959 | 3.15 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr16_+_17269845 | 3.14 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr11_-_69576363 | 3.12 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr7_+_28140352 | 3.08 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr12_-_103322226 | 3.06 |
ENSMUST00000021617.14
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr16_+_78727829 | 3.03 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr17_-_74766131 | 3.03 |
ENSMUST00000052124.9
|
Nlrc4
|
NLR family, CARD domain containing 4 |
| chr6_-_125213911 | 3.01 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr16_+_45430743 | 3.01 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr13_-_4329421 | 3.01 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr13_-_33035150 | 2.99 |
ENSMUST00000091668.13
ENSMUST00000076352.8 |
Serpinb1a
|
serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| chr9_-_104214920 | 2.99 |
ENSMUST00000062723.14
ENSMUST00000215852.2 |
Acpp
|
acid phosphatase, prostate |
| chr19_-_57185988 | 2.99 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr1_+_135727571 | 2.98 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr8_+_46338557 | 2.98 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr15_-_75620060 | 2.94 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr9_-_37166699 | 2.92 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr17_-_34170401 | 2.90 |
ENSMUST00000087543.5
|
B3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr7_+_28140450 | 2.88 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr9_-_66951025 | 2.87 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr5_+_31274064 | 2.87 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr15_+_61858883 | 2.87 |
ENSMUST00000159338.2
|
Myc
|
myelocytomatosis oncogene |
| chr3_+_98129463 | 2.82 |
ENSMUST00000029469.5
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr4_-_49681954 | 2.81 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr1_+_88154727 | 2.80 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr2_-_118380149 | 2.79 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr6_+_127538268 | 2.79 |
ENSMUST00000212051.2
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr4_+_33062999 | 2.78 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr14_+_30601157 | 2.77 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr15_+_101936615 | 2.75 |
ENSMUST00000023803.8
|
Krt18
|
keratin 18 |
| chr17_+_37180437 | 2.73 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr17_+_34544632 | 2.73 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr9_-_66951151 | 2.71 |
ENSMUST00000113696.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr16_+_57173632 | 2.71 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr5_+_77414031 | 2.68 |
ENSMUST00000113449.2
|
Rest
|
RE1-silencing transcription factor |
| chr7_+_28488380 | 2.68 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr16_-_16377982 | 2.66 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_+_56602461 | 2.65 |
ENSMUST00000007340.4
|
Atp12a
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr10_+_127871493 | 2.62 |
ENSMUST00000092048.13
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr4_-_128154709 | 2.62 |
ENSMUST00000053830.5
|
Hmgb4
|
high-mobility group box 4 |
| chr11_+_81936531 | 2.61 |
ENSMUST00000021011.3
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr5_-_65117375 | 2.61 |
ENSMUST00000062315.7
ENSMUST00000239485.2 ENSMUST00000201307.3 |
Tlr6
|
toll-like receptor 6 |
| chr4_-_4077510 | 2.60 |
ENSMUST00000108383.2
|
Sdr16c6
|
short chain dehydrogenase/reductase family 16C, member 6 |
| chr5_+_137639538 | 2.57 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr6_+_41928559 | 2.56 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr4_-_116228921 | 2.55 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr9_-_66950991 | 2.55 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chrX_-_105528821 | 2.55 |
ENSMUST00000039447.14
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr3_-_75177378 | 2.55 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr12_-_113700190 | 2.52 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr17_-_34218301 | 2.51 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr15_-_100567377 | 2.51 |
ENSMUST00000182814.8
ENSMUST00000238935.2 ENSMUST00000182068.8 ENSMUST00000182574.2 ENSMUST00000182775.8 |
Bin2
|
bridging integrator 2 |
| chr6_+_4505493 | 2.50 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr2_+_84670956 | 2.49 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr1_-_182929025 | 2.48 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr12_+_24758724 | 2.48 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr1_-_53391778 | 2.47 |
ENSMUST00000236737.2
ENSMUST00000027264.10 ENSMUST00000123519.9 |
Gm50478
Asnsd1
|
predicted gene, 50478 asparagine synthetase domain containing 1 |
| chr4_+_46489248 | 2.46 |
ENSMUST00000030018.5
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr7_-_45375205 | 2.45 |
ENSMUST00000094424.7
|
Spaca4
|
sperm acrosome associated 4 |
| chr1_+_135980508 | 2.44 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr16_+_48662894 | 2.42 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr5_-_137246611 | 2.40 |
ENSMUST00000196391.5
|
Muc3a
|
mucin 3A, cell surface associated |
| chr6_-_68994064 | 2.39 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr1_+_135764092 | 2.37 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr1_+_135980488 | 2.37 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr6_+_92846338 | 2.36 |
ENSMUST00000113434.2
|
Gm15737
|
predicted gene 15737 |
| chr2_-_76812799 | 2.34 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr3_-_105708632 | 2.34 |
ENSMUST00000090678.11
|
Rap1a
|
RAS-related protein 1a |
| chr17_+_75772475 | 2.32 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr3_-_116222371 | 2.31 |
ENSMUST00000239145.2
|
Cdc14a
|
CDC14 cell division cycle 14A |
| chr1_+_87254729 | 2.30 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr19_+_5118103 | 2.29 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr1_+_135980639 | 2.29 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr11_+_70548513 | 2.27 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr3_-_105708601 | 2.26 |
ENSMUST00000197094.5
ENSMUST00000198004.2 |
Rap1a
|
RAS-related protein 1a |
| chr12_-_103423472 | 2.26 |
ENSMUST00000044687.7
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr9_-_66951114 | 2.25 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr5_-_137246852 | 2.24 |
ENSMUST00000179412.2
|
Muc3a
|
mucin 3A, cell surface associated |
| chr16_+_93629009 | 2.24 |
ENSMUST00000044068.10
ENSMUST00000202261.5 ENSMUST00000201097.3 |
Morc3
|
microrchidia 3 |
| chr11_+_101221895 | 2.19 |
ENSMUST00000017316.7
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr18_-_20192535 | 2.15 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr1_-_165955445 | 2.14 |
ENSMUST00000085992.4
ENSMUST00000192369.6 |
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr1_+_135727228 | 2.13 |
ENSMUST00000154463.8
ENSMUST00000139986.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr5_-_138169509 | 2.12 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr19_-_57185928 | 2.12 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_80338600 | 2.06 |
ENSMUST00000122255.8
|
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr11_+_115790768 | 2.05 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr7_-_131756447 | 2.04 |
ENSMUST00000033149.5
|
Cpxm2
|
carboxypeptidase X 2 (M14 family) |
| chr7_+_27879650 | 2.03 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr14_+_53903370 | 2.03 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr11_-_31320065 | 2.02 |
ENSMUST00000020546.3
|
Stc2
|
stanniocalcin 2 |
| chr4_-_147894245 | 2.01 |
ENSMUST00000105734.10
ENSMUST00000176201.2 |
Zfp984
Gm20707
|
zinc finger protein 984 predicted gene 20707 |
| chr4_-_155947819 | 2.00 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr15_-_100567412 | 1.99 |
ENSMUST00000183211.8
|
Bin2
|
bridging integrator 2 |
| chr2_-_146927365 | 1.99 |
ENSMUST00000067020.3
|
Nkx2-4
|
NK2 homeobox 4 |
| chr2_+_153334710 | 1.99 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr11_-_48836975 | 1.99 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr15_+_103148824 | 1.98 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr15_-_57998443 | 1.98 |
ENSMUST00000038194.5
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr11_+_106642052 | 1.98 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr2_+_29968596 | 1.97 |
ENSMUST00000045246.8
|
Pkn3
|
protein kinase N3 |
| chr17_+_34573760 | 1.96 |
ENSMUST00000178562.2
ENSMUST00000025198.15 |
Btnl2
|
butyrophilin-like 2 |
| chr19_-_57185808 | 1.96 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_-_133599616 | 1.95 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr2_-_7400780 | 1.93 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr18_+_20691095 | 1.89 |
ENSMUST00000059787.15
ENSMUST00000120102.8 |
Dsg2
|
desmoglein 2 |
| chr11_-_69439934 | 1.88 |
ENSMUST00000108659.2
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr16_+_36514334 | 1.86 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 2.8 | 8.5 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 2.7 | 10.9 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 2.3 | 7.0 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 2.2 | 10.8 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 2.1 | 6.4 | GO:0097021 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.0 | 6.0 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.9 | 11.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.8 | 7.2 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.7 | 6.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.7 | 6.7 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.6 | 4.9 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.6 | 9.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.6 | 4.8 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.6 | 3.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 1.5 | 6.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.2 | 4.9 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.2 | 13.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.2 | 4.7 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 1.1 | 6.8 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.1 | 3.3 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 1.1 | 4.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.1 | 6.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 | 2.8 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.9 | 3.6 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.9 | 2.6 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.8 | 1.7 | GO:0071611 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.8 | 6.6 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.8 | 2.5 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.8 | 5.6 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.8 | 7.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 5.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.7 | 2.9 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.7 | 2.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.7 | 2.7 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.7 | 10.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.6 | 2.6 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.6 | 2.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.6 | 2.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.6 | 3.4 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.6 | 4.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.5 | 3.8 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.5 | 10.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.5 | 3.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 3.1 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.5 | 1.6 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.5 | 4.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 2.5 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.5 | 2.0 | GO:0033380 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.5 | 2.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.5 | 2.3 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.5 | 3.7 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.4 | 3.5 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.4 | 1.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 5.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.4 | 1.7 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.4 | 1.3 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.4 | 1.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 1.6 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.4 | 3.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 | 8.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.4 | 1.6 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.4 | 1.6 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.4 | 1.6 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.4 | 4.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 3.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.4 | 2.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 2.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 4.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 1.1 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.4 | 1.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 2.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.0 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.3 | 6.4 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 1.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.3 | 2.0 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.3 | 1.0 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.3 | 6.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.3 | 8.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.3 | 1.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 1.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.3 | 2.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 1.8 | GO:2000561 | negative regulation of T cell cytokine production(GO:0002725) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.3 | 6.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.2 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.3 | 0.9 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.3 | 1.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 3.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 0.8 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 | 1.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 2.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 0.8 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 0.7 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.2 | 0.5 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.2 | 1.0 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 3.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 1.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 2.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 2.6 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 0.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 2.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.7 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.2 | 0.4 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.2 | 1.3 | GO:0043366 | beta selection(GO:0043366) |
| 0.2 | 2.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 3.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.2 | 1.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.6 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.2 | 0.6 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 1.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 1.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.8 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 1.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 1.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 1.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 1.3 | GO:0071896 | synaptic vesicle targeting(GO:0016080) cochlear nucleus development(GO:0021747) protein localization to adherens junction(GO:0071896) |
| 0.2 | 3.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.2 | 1.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.2 | 3.7 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 12.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 0.7 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 3.8 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.2 | 2.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.5 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 1.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 2.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 1.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.5 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.5 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 0.8 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 6.5 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.2 | 2.3 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 5.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 2.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 2.9 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.6 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 1.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 1.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 0.4 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) |
| 0.1 | 2.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.4 | GO:0048625 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.1 | 2.3 | GO:0060033 | anatomical structure regression(GO:0060033) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 2.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 1.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.9 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 4.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.8 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.8 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.5 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 1.6 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 3.9 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.9 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 1.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.7 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 2.7 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 2.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 2.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 2.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.1 | 0.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.4 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 1.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 8.7 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 22.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.8 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.1 | 3.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.6 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 1.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.7 | GO:0042148 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.1 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 4.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 8.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 2.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 2.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.7 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 4.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 3.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.1 | GO:2000556 | T-helper 1 cell cytokine production(GO:0035744) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.1 | 10.0 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 1.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 3.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.9 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 1.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 3.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 3.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.9 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.6 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.7 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.2 | GO:1902477 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.4 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 1.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 2.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 4.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 9.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.4 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 3.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 1.8 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.2 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.3 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.7 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 1.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 4.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.2 | GO:0043585 | nose morphogenesis(GO:0043585) alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.9 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 1.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.8 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 2.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.5 | GO:0051384 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.8 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 1.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 3.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 2.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.3 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 2.4 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.9 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.5 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 2.2 | 6.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 2.2 | 10.9 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 2.1 | 8.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 1.2 | 3.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.1 | 6.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 2.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.8 | 15.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.8 | 11.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.7 | 11.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 3.0 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.6 | 20.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.6 | 1.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 5.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 3.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 3.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 3.2 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 8.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 3.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 5.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.3 | 11.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 1.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 4.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 16.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.3 | 2.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 21.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.2 | 13.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 5.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 1.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 6.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.4 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 0.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 5.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 0.9 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 2.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 9.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 9.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 2.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 6.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 10.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 5.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 10.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 41.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 7.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 12.7 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 17.0 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 3.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 10.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 8.0 | GO:0005694 | chromosome(GO:0005694) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 2.1 | 6.4 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.8 | 10.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.6 | 4.7 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 1.2 | 4.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.1 | 6.9 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.1 | 3.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 2.8 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.9 | 3.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.9 | 2.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.8 | 11.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 3.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.7 | 2.2 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.7 | 2.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.7 | 2.6 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.7 | 8.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.6 | 6.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 3.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.6 | 2.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.6 | 2.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.5 | 3.6 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.4 | 2.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 5.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 6.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 3.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 1.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 2.9 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.4 | 28.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 5.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 3.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 1.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 7.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 1.2 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.4 | 1.5 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.4 | 5.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.4 | 3.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 6.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 1.3 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.3 | 2.9 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 1.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 6.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 1.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.3 | 1.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 2.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 1.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 2.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 3.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 0.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 1.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 6.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 4.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.2 | 1.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 2.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 8.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 5.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.8 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 1.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 6.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.2 | 0.7 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 1.3 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 5.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 1.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 2.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.6 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 2.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.0 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 1.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.7 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 3.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 4.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 4.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 2.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 3.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.6 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 3.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 7.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 6.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 5.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.4 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 3.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 7.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 4.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.7 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 23.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 4.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 4.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 4.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 5.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 7.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 7.6 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 3.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 9.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 1.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 1.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 6.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 16.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 4.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 8.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 1.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 2.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 19.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 4.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 8.0 | GO:0000975 | regulatory region DNA binding(GO:0000975) |
| 0.0 | 3.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 16.5 | GO:0003677 | DNA binding(GO:0003677) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 4.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 10.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 15.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 12.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 8.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 11.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 7.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 4.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 6.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 5.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 8.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 2.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 2.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 6.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 8.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 8.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 7.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 12.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 9.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 16.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.9 | 11.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 5.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 3.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.4 | 27.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 4.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 6.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 3.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 4.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 8.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 6.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 12.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 5.9 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 7.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 19.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 9.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 8.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 5.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.0 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 5.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.6 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 3.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 4.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 11.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 3.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.6 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |