Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
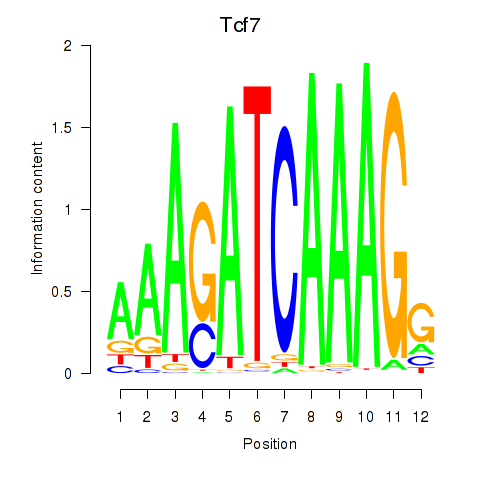
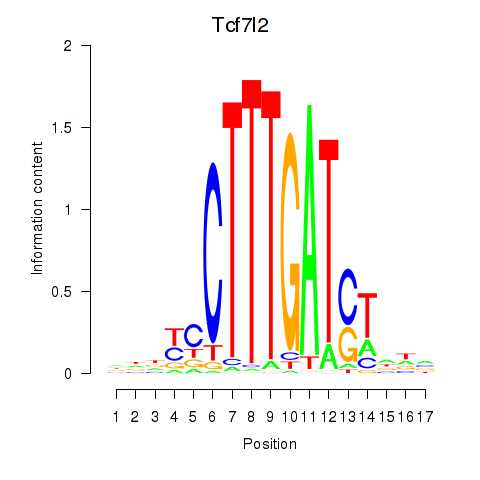
Results for Tcf7_Tcf7l2
Z-value: 1.82
Transcription factors associated with Tcf7_Tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7
|
ENSMUSG00000000782.17 | Tcf7 |
|
Tcf7l2
|
ENSMUSG00000024985.21 | Tcf7l2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7l2 | mm39_v1_chr19_+_55730316_55730351 | 0.19 | 1.1e-01 | Click! |
| Tcf7 | mm39_v1_chr11_-_52173391_52173405 | -0.15 | 1.9e-01 | Click! |
Activity profile of Tcf7_Tcf7l2 motif
Sorted Z-values of Tcf7_Tcf7l2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7_Tcf7l2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_22224506 | 23.53 |
ENSMUST00000080533.6
|
Defa24
|
defensin, alpha, 24 |
| chr8_+_21691577 | 23.10 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr8_+_21787455 | 21.51 |
ENSMUST00000098892.5
|
Defa5
|
defensin, alpha, 5 |
| chr8_+_22155813 | 19.03 |
ENSMUST00000075268.5
|
Defa34
|
defensin, alpha, 34 |
| chr3_-_144638284 | 18.25 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr3_+_57332735 | 16.01 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr2_+_121188195 | 14.73 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr9_+_46151994 | 14.62 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr5_-_145816774 | 14.46 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr4_+_11758147 | 14.44 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr9_+_7692087 | 13.31 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
| chr18_+_21205386 | 12.85 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr13_+_54849268 | 10.98 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr8_+_21652293 | 10.93 |
ENSMUST00000098897.2
|
Defa22
|
defensin, alpha, 22 |
| chr1_+_88093726 | 10.77 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr9_+_98368163 | 10.75 |
ENSMUST00000189446.7
ENSMUST00000187905.7 |
Rbp2
|
retinol binding protein 2, cellular |
| chr8_+_21868531 | 10.74 |
ENSMUST00000170275.4
|
Defa2
|
defensin, alpha, 2 |
| chr8_+_21881827 | 10.39 |
ENSMUST00000120874.5
|
Defa33
|
defensin, alpha, 33 |
| chr8_+_21805562 | 10.24 |
ENSMUST00000167683.3
ENSMUST00000168340.2 |
Defa27
|
defensin, alpha, 27 |
| chr3_-_144680801 | 10.23 |
ENSMUST00000029923.10
ENSMUST00000238960.2 |
Clca4a
|
chloride channel accessory 4A |
| chr12_-_114487525 | 10.18 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr8_+_21555054 | 10.04 |
ENSMUST00000078121.4
|
Defa35
|
defensin, alpha, 35 |
| chr18_+_35860019 | 9.52 |
ENSMUST00000097617.3
|
1700066B19Rik
|
RIKEN cDNA 1700066B19 gene |
| chr19_+_44980565 | 9.32 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr1_+_157334347 | 9.04 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334298 | 8.83 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr14_+_56255422 | 7.99 |
ENSMUST00000022836.6
|
Mcpt1
|
mast cell protease 1 |
| chr8_+_22019048 | 7.62 |
ENSMUST00000084041.4
|
Defa32
|
defensin, alpha, 32 |
| chr13_-_100338469 | 7.57 |
ENSMUST00000167986.3
ENSMUST00000117913.8 |
Naip2
|
NLR family, apoptosis inhibitory protein 2 |
| chr8_+_4399588 | 7.48 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr12_-_76842263 | 7.29 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr8_+_22073022 | 7.22 |
ENSMUST00000098887.4
|
Defa28
|
defensin, alpha, 28 |
| chr11_+_96820091 | 6.81 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr5_-_87716882 | 6.68 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr3_+_118355778 | 6.58 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr7_-_30776081 | 6.51 |
ENSMUST00000072331.13
ENSMUST00000167369.8 |
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr8_+_21515561 | 6.45 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr2_-_154916367 | 6.23 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr2_-_62313981 | 6.04 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr3_+_118355811 | 6.02 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr8_+_22055402 | 5.98 |
ENSMUST00000084040.3
|
Defa37
|
defensin, alpha, 37 |
| chr5_-_73789764 | 5.91 |
ENSMUST00000087177.4
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr11_+_102036356 | 5.78 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr6_-_69800923 | 5.76 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr2_+_163348728 | 5.58 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr3_+_92874366 | 5.55 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr12_-_113790741 | 5.14 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr7_-_79492091 | 5.14 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr12_-_115876396 | 4.82 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr1_-_86039692 | 4.77 |
ENSMUST00000027431.7
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr14_-_54651442 | 4.75 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr3_-_92627651 | 4.61 |
ENSMUST00000047153.4
|
Lce1f
|
late cornified envelope 1F |
| chr11_+_96820220 | 4.56 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr2_-_127571836 | 4.54 |
ENSMUST00000028856.3
|
Mall
|
mal, T cell differentiation protein-like |
| chr4_+_45848918 | 4.49 |
ENSMUST00000030011.6
|
Stra6l
|
STRA6-like |
| chr8_+_21739729 | 4.44 |
ENSMUST00000098895.3
|
Defa40
|
defensin, alpha, 40 |
| chr10_-_115423644 | 4.43 |
ENSMUST00000020350.15
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr5_-_145406533 | 4.42 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr8_+_21574411 | 4.41 |
ENSMUST00000095425.2
|
Defa25
|
defensin, alpha, 25 |
| chr10_+_61484331 | 4.40 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr19_+_39049442 | 4.39 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr15_-_54141816 | 4.37 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr6_-_47790272 | 4.21 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr11_+_98337655 | 4.21 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr6_+_68495964 | 4.18 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr10_-_127206300 | 4.17 |
ENSMUST00000026472.10
|
Inhbc
|
inhibin beta-C |
| chr7_+_24310738 | 4.07 |
ENSMUST00000073325.6
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr3_-_92594516 | 4.05 |
ENSMUST00000029524.4
|
Lce1d
|
late cornified envelope 1D |
| chr9_+_45014092 | 4.05 |
ENSMUST00000217074.2
|
Jaml
|
junction adhesion molecule like |
| chr13_-_41981812 | 3.93 |
ENSMUST00000223337.2
ENSMUST00000221691.2 |
Adtrp
|
androgen dependent TFPI regulating protein |
| chr11_-_43492367 | 3.92 |
ENSMUST00000020672.5
|
Fabp6
|
fatty acid binding protein 6 |
| chr13_-_41981893 | 3.81 |
ENSMUST00000137905.2
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr18_+_36797113 | 3.77 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr10_-_24803336 | 3.73 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr12_-_114140482 | 3.70 |
ENSMUST00000103475.2
ENSMUST00000195706.2 |
Ighv14-4
|
immunoglobulin heavy variable 14-4 |
| chr11_+_108811626 | 3.68 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr14_-_34077340 | 3.55 |
ENSMUST00000052126.6
|
Fam25c
|
family with sequence similarity 25, member C |
| chr10_-_128425519 | 3.52 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr10_+_71183571 | 3.39 |
ENSMUST00000079252.13
|
Ipmk
|
inositol polyphosphate multikinase |
| chr1_+_87998487 | 3.25 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr8_-_110688716 | 3.19 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr6_-_69377328 | 3.17 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr2_+_111205867 | 3.09 |
ENSMUST00000062407.6
|
Olfr1284
|
olfactory receptor 1284 |
| chr11_+_58311921 | 3.07 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr14_-_70561231 | 3.05 |
ENSMUST00000151011.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr6_+_34722887 | 2.93 |
ENSMUST00000123823.8
ENSMUST00000136907.8 |
Cald1
|
caldesmon 1 |
| chr16_+_22739028 | 2.92 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr12_-_113802603 | 2.90 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr6_-_69835868 | 2.85 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr15_+_31224460 | 2.81 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr11_+_108812474 | 2.81 |
ENSMUST00000144511.2
|
Axin2
|
axin 2 |
| chr3_-_92659644 | 2.77 |
ENSMUST00000029527.6
|
Lce1g
|
late cornified envelope 1G |
| chr5_-_101812862 | 2.72 |
ENSMUST00000044125.11
|
Nkx6-1
|
NK6 homeobox 1 |
| chr3_-_92686198 | 2.65 |
ENSMUST00000090866.2
|
Lce1i
|
late cornified envelope 1I |
| chr4_-_43499608 | 2.63 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr11_-_100139728 | 2.58 |
ENSMUST00000007280.9
|
Krt16
|
keratin 16 |
| chr6_+_34722926 | 2.58 |
ENSMUST00000126181.8
|
Cald1
|
caldesmon 1 |
| chr1_-_59276252 | 2.51 |
ENSMUST00000163058.2
ENSMUST00000160945.2 ENSMUST00000027178.13 |
Als2
|
alsin Rho guanine nucleotide exchange factor |
| chr1_-_139487951 | 2.49 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr4_-_63072367 | 2.47 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr11_-_86964881 | 2.43 |
ENSMUST00000020804.8
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr2_-_143704853 | 2.42 |
ENSMUST00000099296.4
|
Bfsp1
|
beaded filament structural protein 1, in lens-CP94 |
| chr12_-_114321838 | 2.41 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr16_-_45313324 | 2.41 |
ENSMUST00000114585.3
|
Gm609
|
predicted gene 609 |
| chr1_+_106866678 | 2.40 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr3_-_96721829 | 2.37 |
ENSMUST00000047702.8
|
Cd160
|
CD160 antigen |
| chr10_-_75634407 | 2.36 |
ENSMUST00000001713.10
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr3_-_92672367 | 2.31 |
ENSMUST00000051521.5
|
Lce1h
|
late cornified envelope 1H |
| chr15_+_84926909 | 2.25 |
ENSMUST00000229203.2
|
Fam118a
|
family with sequence similarity 118, member A |
| chr10_-_60983438 | 2.24 |
ENSMUST00000092498.12
ENSMUST00000137833.2 ENSMUST00000155919.8 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr9_-_50657800 | 2.21 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr12_-_84455764 | 2.17 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr5_-_145521533 | 2.17 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr2_-_60213575 | 2.16 |
ENSMUST00000028362.9
|
Ly75
|
lymphocyte antigen 75 |
| chr6_+_138117519 | 2.13 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr9_-_26834356 | 2.12 |
ENSMUST00000115261.3
|
Gm1110
|
predicted gene 1110 |
| chr6_+_68402550 | 2.09 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr8_+_10299288 | 2.08 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr2_-_60213639 | 2.05 |
ENSMUST00000112533.8
|
Ly75
|
lymphocyte antigen 75 |
| chr10_+_75242745 | 2.05 |
ENSMUST00000039925.8
|
Upb1
|
ureidopropionase, beta |
| chr3_+_122305819 | 2.04 |
ENSMUST00000199344.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr6_+_138117295 | 2.04 |
ENSMUST00000008684.11
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr11_+_99755302 | 2.03 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr3_+_92840279 | 2.00 |
ENSMUST00000046234.5
|
Lce3b
|
late cornified envelope 3B |
| chr11_-_86999481 | 1.99 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr17_-_35077089 | 1.97 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr13_-_56696310 | 1.96 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr13_-_104056803 | 1.96 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr2_+_70305267 | 1.92 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr1_+_87983099 | 1.91 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_+_75139295 | 1.91 |
ENSMUST00000036555.8
|
Myo5c
|
myosin VC |
| chr19_+_56276343 | 1.90 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr10_+_18283405 | 1.88 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr3_-_116506345 | 1.86 |
ENSMUST00000169530.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr13_+_54738014 | 1.85 |
ENSMUST00000026986.7
|
Higd2a
|
HIG1 domain family, member 2A |
| chr16_-_10884005 | 1.83 |
ENSMUST00000162323.2
|
Litaf
|
LPS-induced TN factor |
| chr11_-_59927688 | 1.83 |
ENSMUST00000102692.10
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr2_+_152873772 | 1.82 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr10_+_101517348 | 1.82 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr3_+_92586546 | 1.78 |
ENSMUST00000047055.4
|
Lce1c
|
late cornified envelope 1C |
| chr5_-_44259374 | 1.78 |
ENSMUST00000171543.8
|
Prom1
|
prominin 1 |
| chrX_+_59044796 | 1.77 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr11_-_16458069 | 1.76 |
ENSMUST00000109641.2
|
Sec61g
|
SEC61, gamma subunit |
| chr11_-_16458148 | 1.74 |
ENSMUST00000109642.8
|
Sec61g
|
SEC61, gamma subunit |
| chr3_+_92864693 | 1.74 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr6_-_69584812 | 1.71 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr6_-_69753317 | 1.70 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr19_-_39875192 | 1.68 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr18_+_84106188 | 1.68 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr1_+_87983189 | 1.68 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr14_+_54374195 | 1.67 |
ENSMUST00000103683.2
|
Trdj2
|
T cell receptor delta joining 2 |
| chr13_+_94219934 | 1.67 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr16_-_56533179 | 1.65 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr15_+_85743887 | 1.64 |
ENSMUST00000170629.3
|
Gtse1
|
G two S phase expressed protein 1 |
| chr8_-_107792264 | 1.61 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_38816229 | 1.61 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr13_-_56696222 | 1.61 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr10_+_85763545 | 1.61 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr6_+_54016543 | 1.59 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr7_+_25760922 | 1.59 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr10_+_126836578 | 1.58 |
ENSMUST00000026500.12
ENSMUST00000142698.8 |
Avil
|
advillin |
| chr10_+_42736771 | 1.57 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr4_+_95855442 | 1.56 |
ENSMUST00000030306.14
|
Hook1
|
hook microtubule tethering protein 1 |
| chr16_+_87495792 | 1.54 |
ENSMUST00000026703.6
|
Bach1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr3_-_53771185 | 1.53 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr7_+_24310171 | 1.52 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr15_+_85744147 | 1.51 |
ENSMUST00000231074.2
|
Gtse1
|
G two S phase expressed protein 1 |
| chr3_+_85946145 | 1.50 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr13_+_19394484 | 1.50 |
ENSMUST00000200495.2
|
Trgj1
|
T cell receptor gamma joining 1 |
| chr18_+_61688329 | 1.49 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr6_-_52211882 | 1.48 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr1_+_131566044 | 1.48 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr9_+_21527462 | 1.48 |
ENSMUST00000034707.15
ENSMUST00000098948.10 |
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr7_-_44711075 | 1.48 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr5_-_113217983 | 1.46 |
ENSMUST00000112336.8
|
Crybb2
|
crystallin, beta B2 |
| chr11_+_108271990 | 1.45 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr2_-_151474391 | 1.44 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr5_-_44259293 | 1.42 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr19_+_5928649 | 1.40 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr7_-_44711130 | 1.39 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr10_-_18890281 | 1.39 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr9_-_15212745 | 1.36 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr2_-_30095784 | 1.35 |
ENSMUST00000113662.8
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr17_-_34822649 | 1.33 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr4_+_95855560 | 1.33 |
ENSMUST00000107083.2
|
Hook1
|
hook microtubule tethering protein 1 |
| chr8_-_55340024 | 1.32 |
ENSMUST00000176866.8
|
Wdr17
|
WD repeat domain 17 |
| chr12_-_113823290 | 1.32 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr13_-_23015518 | 1.32 |
ENSMUST00000226294.2
ENSMUST00000226180.2 |
Vmn1r210
|
vomeronasal 1 receptor 210 |
| chr11_+_108814007 | 1.31 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr6_-_129761233 | 1.30 |
ENSMUST00000118532.8
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr10_+_80971054 | 1.30 |
ENSMUST00000125261.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr16_-_45313244 | 1.28 |
ENSMUST00000232138.2
|
Gm609
|
predicted gene 609 |
| chr7_+_79675727 | 1.28 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr14_+_102078038 | 1.28 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr9_+_65967477 | 1.27 |
ENSMUST00000034947.7
|
Ppib
|
peptidylprolyl isomerase B |
| chr5_+_18167547 | 1.27 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr15_-_11399666 | 1.23 |
ENSMUST00000022849.7
|
Tars
|
threonyl-tRNA synthetase |
| chr11_-_99412084 | 1.22 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr19_+_55882942 | 1.22 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr7_+_79676095 | 1.21 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.6 | GO:0044240 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 4.8 | 14.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 4.4 | 13.3 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 4.2 | 12.6 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 2.2 | 11.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.9 | 5.6 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.8 | 7.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.8 | 17.9 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 1.6 | 6.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 1.5 | 6.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.5 | 4.5 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 1.5 | 17.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.4 | 4.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 1.3 | 7.8 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.2 | 10.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.2 | 4.8 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 1.0 | 4.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.0 | 6.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.8 | 2.5 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.8 | 9.5 | GO:0000050 | urea cycle(GO:0000050) |
| 0.8 | 2.4 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 0.8 | 6.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.7 | 23.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.6 | 3.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.6 | 8.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.6 | 4.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 3.3 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.5 | 2.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.5 | 1.0 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.5 | 3.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.5 | 4.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 4.4 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.4 | 1.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.4 | 27.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 1.9 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.4 | 1.1 | GO:2000813 | actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.4 | 1.4 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.3 | 1.5 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 1.5 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 9.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 1.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 2.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 0.7 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.2 | 1.5 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.2 | 5.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.2 | 2.2 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.2 | 0.9 | GO:0050823 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.9 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 0.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 17.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 13.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 0.6 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.2 | 1.3 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.8 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 1.1 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.6 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.2 | 1.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 30.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 16.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.2 | 1.8 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 0.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 50.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.2 | 4.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.7 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.2 | 1.6 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 3.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 2.6 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 1.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 2.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 3.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 0.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.2 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 3.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 4.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 2.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 2.2 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.2 | 0.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.2 | 3.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 2.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.6 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 1.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.4 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 1.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 1.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.5 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 2.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.5 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 1.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 4.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 0.7 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 1.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.8 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 5.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 6.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.6 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 1.0 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 1.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 3.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.8 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 12.3 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 0.6 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 4.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.0 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.8 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 4.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.5 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 3.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 2.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 17.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.8 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.1 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 2.4 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.8 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 2.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.5 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.8 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.6 | GO:0019243 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 4.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 1.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.7 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 1.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 3.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 3.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 1.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 1.1 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.5 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.1 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 1.4 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 1.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.7 | GO:0050913 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.5 | GO:0060065 | proximal/distal pattern formation(GO:0009954) uterus development(GO:0060065) |
| 0.0 | 2.7 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.5 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 2.1 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.3 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 3.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.0 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.1 | 7.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.7 | 16.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.6 | 5.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 26.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 5.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.5 | 1.8 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.5 | 3.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 2.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 5.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 30.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 1.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.9 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 1.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 2.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 4.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.6 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 11.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 8.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 133.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 4.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 4.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 2.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 6.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 4.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 4.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 13.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 35.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 3.6 | 14.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 2.6 | 21.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 2.2 | 4.4 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 2.1 | 14.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.9 | 7.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.9 | 5.6 | GO:0070540 | stearic acid binding(GO:0070540) |
| 1.8 | 14.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.3 | 6.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.1 | 3.4 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 1.1 | 4.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 1.0 | 5.8 | GO:0034618 | arginine binding(GO:0034618) |
| 0.9 | 28.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.9 | 6.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.8 | 2.5 | GO:0047945 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.8 | 2.4 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.6 | 2.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 10.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 2.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.4 | 17.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.7 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 1.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 1.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.4 | 3.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 3.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 7.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.4 | 11.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 1.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.4 | 1.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 3.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 1.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.3 | 4.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 1.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.3 | 4.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 0.8 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.3 | 2.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 4.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 4.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 30.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.6 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 0.6 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.2 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 3.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 5.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 2.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 2.8 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 3.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.8 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.2 | 1.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 21.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 5.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 5.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 3.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.5 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.9 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.4 | GO:0016420 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.4 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 3.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 5.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.9 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 6.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 7.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 10.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.9 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 15.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.6 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.5 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.1 | 1.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 4.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 4.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 14.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 18.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.0 | GO:0048029 | monosaccharide binding(GO:0048029) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 23.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 4.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 8.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 6.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 8.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 4.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 6.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 20.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.3 | 14.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 14.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 14.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 5.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.4 | 4.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 3.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 6.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 6.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 6.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 5.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 1.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 4.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 5.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 4.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.9 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 3.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |