Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
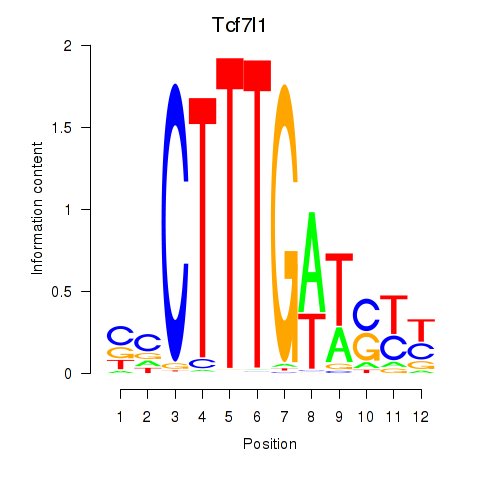
Results for Tcf7l1
Z-value: 0.83
Transcription factors associated with Tcf7l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7l1
|
ENSMUSG00000055799.14 | Tcf7l1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7l1 | mm39_v1_chr6_-_72765935_72766044 | 0.34 | 3.7e-03 | Click! |
Activity profile of Tcf7l1 motif
Sorted Z-values of Tcf7l1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7l1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76842263 | 6.84 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr5_-_134975773 | 5.13 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr15_+_102898966 | 4.55 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chrX_+_72527208 | 4.54 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chr11_+_96820220 | 4.39 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr6_-_83504471 | 4.39 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr11_+_96820091 | 4.01 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr18_+_21205386 | 3.99 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr10_+_79986280 | 3.75 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chr17_-_71158184 | 3.37 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_-_69416365 | 3.26 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr3_-_144638284 | 3.25 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr6_+_21985902 | 3.23 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_-_71158052 | 3.22 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr12_+_113104085 | 3.20 |
ENSMUST00000200380.5
|
Crip2
|
cysteine rich protein 2 |
| chr3_-_92594516 | 3.16 |
ENSMUST00000029524.4
|
Lce1d
|
late cornified envelope 1D |
| chr4_+_141148068 | 3.12 |
ENSMUST00000102486.5
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr2_-_65068960 | 3.11 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr2_-_65068917 | 3.11 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr9_+_98372575 | 3.10 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr8_+_21691577 | 3.05 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr4_+_11758147 | 3.02 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr12_+_113103817 | 3.02 |
ENSMUST00000084882.9
|
Crip2
|
cysteine rich protein 2 |
| chr6_-_52211882 | 2.97 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr14_+_101967343 | 2.93 |
ENSMUST00000100337.10
|
Lmo7
|
LIM domain only 7 |
| chr6_+_17065141 | 2.89 |
ENSMUST00000115467.11
ENSMUST00000154266.3 ENSMUST00000076654.9 |
Tes
|
testin LIM domain protein |
| chr4_-_94538329 | 2.86 |
ENSMUST00000107101.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr2_+_70305267 | 2.80 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chrX_+_162923474 | 2.71 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr10_+_38841511 | 2.70 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr11_-_52174129 | 2.66 |
ENSMUST00000109071.3
|
Tcf7
|
transcription factor 7, T cell specific |
| chr17_+_75312520 | 2.64 |
ENSMUST00000234490.2
ENSMUST00000001927.12 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_68311073 | 2.61 |
ENSMUST00000024840.12
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr10_-_85847697 | 2.59 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr6_+_21986445 | 2.59 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr11_+_108812474 | 2.45 |
ENSMUST00000144511.2
|
Axin2
|
axin 2 |
| chr3_+_122688721 | 2.43 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr3_-_92555608 | 2.42 |
ENSMUST00000074142.7
|
Lce1a1
|
late cornified envelope 1A1 |
| chr3_-_75864195 | 2.42 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr1_-_181847492 | 2.36 |
ENSMUST00000177811.8
ENSMUST00000111025.8 ENSMUST00000111024.10 |
Enah
|
ENAH actin regulator |
| chr6_-_148846247 | 2.34 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr18_+_4921663 | 2.32 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chr12_-_84455764 | 2.31 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr12_-_112766266 | 2.30 |
ENSMUST00000239525.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr12_-_16850674 | 2.25 |
ENSMUST00000162112.8
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr3_-_92616380 | 2.24 |
ENSMUST00000090867.2
|
Lce1e
|
late cornified envelope 1E |
| chr14_+_34097474 | 2.24 |
ENSMUST00000227130.2
|
Mmrn2
|
multimerin 2 |
| chr11_+_108811626 | 2.19 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr4_-_94538370 | 2.19 |
ENSMUST00000053419.9
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr2_+_74535242 | 2.14 |
ENSMUST00000019749.4
|
Hoxd8
|
homeobox D8 |
| chr7_+_43460718 | 2.11 |
ENSMUST00000032955.7
|
Klk7
|
kallikrein related-peptidase 7 (chymotryptic, stratum corneum) |
| chr2_-_35322581 | 2.09 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr9_+_90045219 | 2.09 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_+_90045109 | 2.08 |
ENSMUST00000113059.8
ENSMUST00000167122.8 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr14_+_34097422 | 2.07 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr6_+_29735666 | 2.06 |
ENSMUST00000001812.5
|
Smo
|
smoothened, frizzled class receptor |
| chr8_+_79755194 | 2.04 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr2_+_174602412 | 1.99 |
ENSMUST00000029030.9
|
Edn3
|
endothelin 3 |
| chr13_-_119545479 | 1.99 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr11_-_48836975 | 1.99 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr8_+_93084253 | 1.98 |
ENSMUST00000210246.2
ENSMUST00000034184.12 |
Irx5
|
Iroquois homeobox 5 |
| chr15_-_54141816 | 1.95 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr14_+_102078038 | 1.93 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr11_-_53959790 | 1.89 |
ENSMUST00000018755.10
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr11_-_53959758 | 1.89 |
ENSMUST00000093109.11
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr13_-_119545520 | 1.86 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr7_-_144493560 | 1.86 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr8_+_79754980 | 1.83 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr1_-_86039692 | 1.82 |
ENSMUST00000027431.7
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr6_+_83034825 | 1.81 |
ENSMUST00000077502.5
|
Dqx1
|
DEAQ RNA-dependent ATPase |
| chr6_-_47571901 | 1.80 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr6_-_47790272 | 1.78 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr7_+_88079534 | 1.78 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr19_-_47907705 | 1.77 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr11_-_86999481 | 1.77 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr10_-_4337435 | 1.73 |
ENSMUST00000100077.5
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr1_-_37996838 | 1.69 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr2_+_74534959 | 1.69 |
ENSMUST00000151380.2
|
Hoxd8
|
homeobox D8 |
| chr10_+_96453408 | 1.69 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr17_+_37504783 | 1.68 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr17_+_29712008 | 1.67 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr10_+_79986988 | 1.66 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chr10_+_126836578 | 1.64 |
ENSMUST00000026500.12
ENSMUST00000142698.8 |
Avil
|
advillin |
| chr3_+_92874366 | 1.60 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr10_-_4338032 | 1.59 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr14_+_26760898 | 1.59 |
ENSMUST00000035336.5
|
Il17rd
|
interleukin 17 receptor D |
| chr4_-_34882917 | 1.58 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr14_-_70761507 | 1.56 |
ENSMUST00000022692.5
|
Sftpc
|
surfactant associated protein C |
| chr15_-_58078274 | 1.56 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chrX_-_133062677 | 1.51 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr2_+_121188195 | 1.51 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr13_-_103042554 | 1.49 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_-_103042294 | 1.49 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_-_147244074 | 1.47 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr11_+_120382666 | 1.46 |
ENSMUST00000026899.4
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr14_-_98406977 | 1.46 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr6_+_86605146 | 1.41 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr3_+_88439616 | 1.40 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr10_-_13264497 | 1.40 |
ENSMUST00000105546.8
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr8_+_21805562 | 1.40 |
ENSMUST00000167683.3
ENSMUST00000168340.2 |
Defa27
|
defensin, alpha, 27 |
| chr10_-_61814852 | 1.39 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr1_+_107289659 | 1.38 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr14_+_26761023 | 1.37 |
ENSMUST00000223942.2
|
Il17rd
|
interleukin 17 receptor D |
| chr11_+_95010935 | 1.37 |
ENSMUST00000092768.7
|
Dlx3
|
distal-less homeobox 3 |
| chr11_-_86964881 | 1.36 |
ENSMUST00000020804.8
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr9_-_57552392 | 1.34 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr1_-_182929025 | 1.34 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr8_+_22996233 | 1.33 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr12_-_114487525 | 1.33 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr7_+_24310738 | 1.32 |
ENSMUST00000073325.6
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr18_-_61169262 | 1.32 |
ENSMUST00000025521.9
|
Cdx1
|
caudal type homeobox 1 |
| chr11_-_49603501 | 1.31 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr12_-_21467437 | 1.31 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr8_+_61677253 | 1.30 |
ENSMUST00000209611.2
|
Sh3rf1
|
SH3 domain containing ring finger 1 |
| chr2_+_174602574 | 1.30 |
ENSMUST00000140908.2
|
Edn3
|
endothelin 3 |
| chr4_+_62501737 | 1.28 |
ENSMUST00000098031.10
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr12_+_76118790 | 1.28 |
ENSMUST00000131480.8
|
Syne2
|
spectrin repeat containing, nuclear envelope 2 |
| chr12_-_98703664 | 1.28 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr9_+_45014092 | 1.26 |
ENSMUST00000217074.2
|
Jaml
|
junction adhesion molecule like |
| chr7_+_78922947 | 1.26 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr2_+_172863688 | 1.24 |
ENSMUST00000029014.16
|
Rbm38
|
RNA binding motif protein 38 |
| chr1_+_39940189 | 1.23 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_+_32763313 | 1.23 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr2_-_143704853 | 1.23 |
ENSMUST00000099296.4
|
Bfsp1
|
beaded filament structural protein 1, in lens-CP94 |
| chr17_+_48037758 | 1.21 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr3_-_122413361 | 1.21 |
ENSMUST00000239148.2
ENSMUST00000162409.8 ENSMUST00000162947.3 |
Fnbp1l
|
formin binding protein 1-like |
| chr11_+_108811168 | 1.17 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr11_+_108814007 | 1.16 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr6_+_87350292 | 1.14 |
ENSMUST00000032128.6
|
Gkn2
|
gastrokine 2 |
| chr17_-_79328157 | 1.14 |
ENSMUST00000168887.8
ENSMUST00000119284.8 |
Prkd3
|
protein kinase D3 |
| chr1_-_64161415 | 1.13 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_94219934 | 1.11 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr7_+_5018453 | 1.10 |
ENSMUST00000086349.5
ENSMUST00000207050.2 |
Zfp524
Gm44973
|
zinc finger protein 524 predicted gene 44973 |
| chr17_+_36176485 | 1.09 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr2_+_13579092 | 1.09 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr4_+_57782150 | 1.08 |
ENSMUST00000124581.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr14_+_102077937 | 1.07 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr14_-_55204023 | 1.06 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chrX_+_158771429 | 1.05 |
ENSMUST00000033665.9
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr1_+_136395673 | 1.05 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr9_+_35334878 | 1.04 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr14_+_21549798 | 1.03 |
ENSMUST00000182855.8
ENSMUST00000182405.9 ENSMUST00000069648.14 |
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr13_-_103233323 | 1.01 |
ENSMUST00000166336.9
ENSMUST00000239261.2 ENSMUST00000194446.7 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_25214322 | 1.01 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr19_+_56276343 | 1.00 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr7_+_44866095 | 1.00 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr3_+_32762656 | 1.00 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A |
| chr11_-_61610772 | 0.98 |
ENSMUST00000151780.8
ENSMUST00000148584.2 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr18_-_88912446 | 0.98 |
ENSMUST00000070116.12
ENSMUST00000125362.8 |
Socs6
|
suppressor of cytokine signaling 6 |
| chr17_+_36176948 | 0.95 |
ENSMUST00000122899.8
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr2_+_156928962 | 0.95 |
ENSMUST00000166140.8
|
Tldc2
|
TBC/LysM associated domain containing 2 |
| chr10_-_13264574 | 0.94 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr9_-_57552844 | 0.94 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr6_-_52195663 | 0.93 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr4_-_58553553 | 0.92 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr16_+_87495792 | 0.92 |
ENSMUST00000026703.6
|
Bach1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr13_+_22129246 | 0.89 |
ENSMUST00000176511.8
ENSMUST00000102978.8 ENSMUST00000152258.9 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr13_-_99062882 | 0.88 |
ENSMUST00000109401.8
|
Tnpo1
|
transportin 1 |
| chr7_+_98484311 | 0.88 |
ENSMUST00000165122.8
ENSMUST00000067495.9 |
Wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr9_-_65734826 | 0.87 |
ENSMUST00000159109.2
|
Zfp609
|
zinc finger protein 609 |
| chr3_-_116762476 | 0.87 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr14_-_70414236 | 0.87 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr2_-_59955995 | 0.86 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_-_126362372 | 0.85 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr8_+_22073022 | 0.85 |
ENSMUST00000098887.4
|
Defa28
|
defensin, alpha, 28 |
| chr7_-_110681402 | 0.85 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr17_+_35295849 | 0.81 |
ENSMUST00000172494.8
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr7_+_65759198 | 0.81 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chrX_-_149595711 | 0.81 |
ENSMUST00000112697.10
|
Maged2
|
MAGE family member D2 |
| chr2_+_18677195 | 0.79 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr7_-_49286594 | 0.78 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr2_+_160573604 | 0.78 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr11_+_69737200 | 0.76 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chr18_+_82932747 | 0.76 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr12_-_31401432 | 0.75 |
ENSMUST00000110857.5
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr19_-_4384029 | 0.74 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr8_-_41507808 | 0.72 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_+_172864153 | 0.72 |
ENSMUST00000173997.2
|
Rbm38
|
RNA binding motif protein 38 |
| chr19_-_41790458 | 0.72 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr7_+_67305162 | 0.72 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr18_+_61688329 | 0.71 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr18_-_88912322 | 0.71 |
ENSMUST00000123826.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr1_-_4479445 | 0.71 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr1_-_58735106 | 0.70 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chr15_-_37008011 | 0.70 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr11_+_114742619 | 0.69 |
ENSMUST00000053361.12
ENSMUST00000021071.14 ENSMUST00000136785.2 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr12_-_51876282 | 0.69 |
ENSMUST00000042052.9
ENSMUST00000179265.8 |
Hectd1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chr12_+_76128405 | 0.68 |
ENSMUST00000154509.8
|
Syne2
|
spectrin repeat containing, nuclear envelope 2 |
| chr7_+_126359869 | 0.68 |
ENSMUST00000206272.2
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr2_+_152873772 | 0.66 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr1_-_180524587 | 0.65 |
ENSMUST00000027778.8
|
Mixl1
|
Mix1 homeobox-like 1 (Xenopus laevis) |
| chr17_-_46513499 | 0.63 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chrX_-_149595873 | 0.62 |
ENSMUST00000131241.2
ENSMUST00000147152.3 ENSMUST00000143843.8 |
Maged2
|
MAGE family member D2 |
| chr5_+_31350566 | 0.62 |
ENSMUST00000031029.15
ENSMUST00000201679.4 |
Snx17
|
sorting nexin 17 |
| chr17_-_34822649 | 0.62 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr19_+_44481901 | 0.62 |
ENSMUST00000041163.5
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr5_+_36050663 | 0.61 |
ENSMUST00000064571.11
|
Afap1
|
actin filament associated protein 1 |
| chr5_+_31350607 | 0.61 |
ENSMUST00000201535.4
|
Snx17
|
sorting nexin 17 |
| chr1_-_37580084 | 0.61 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr11_-_99501015 | 0.60 |
ENSMUST00000076478.2
|
Gm11937
|
predicted gene 11937 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.3 | 3.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.2 | 7.0 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.0 | 3.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.7 | 5.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.7 | 2.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.7 | 3.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.7 | 2.6 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.6 | 1.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.6 | 1.8 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.5 | 3.3 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.5 | 2.1 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.5 | 1.6 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 2.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.5 | 1.8 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.4 | 1.3 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.4 | 2.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.4 | 1.5 | GO:0015744 | succinate transport(GO:0015744) |
| 0.3 | 1.0 | GO:0032487 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.3 | 3.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.4 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 1.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 2.7 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.3 | 1.3 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 2.3 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.3 | 6.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 4.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 1.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.3 | 1.1 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.3 | 1.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 1.8 | GO:0060155 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.2 | 1.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 1.0 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 2.8 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 6.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.9 | GO:0060775 | positive regulation of transforming growth factor beta2 production(GO:0032915) mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.2 | 5.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 1.7 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.2 | 1.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 0.8 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 2.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.2 | 0.7 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.9 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 1.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 1.7 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.2 | 2.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 1.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 11.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 4.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 2.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.3 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.1 | 0.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 1.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 2.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 2.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.4 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 2.0 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 0.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 3.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.0 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 2.3 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.1 | 3.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.3 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.1 | 0.8 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.7 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 1.0 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 2.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.9 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 1.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 1.0 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 1.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.6 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 4.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 4.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) neutrophil differentiation(GO:0030223) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 2.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 1.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 3.9 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 1.0 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 3.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0003285 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.4 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 1.8 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.6 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 6.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.9 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 2.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.5 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.5 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.4 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.6 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.6 | GO:0042461 | photoreceptor cell development(GO:0042461) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.4 | 2.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 1.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 2.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.7 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 9.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 5.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 1.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 3.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 4.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.3 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 4.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 3.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 4.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 3.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 2.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 9.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 3.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 6.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.6 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.8 | 3.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.8 | 3.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.8 | 3.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.6 | 1.8 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.5 | 1.5 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 2.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.5 | 2.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 2.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 2.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 7.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 3.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.3 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 6.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 2.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.3 | 0.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 1.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 1.8 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 6.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 2.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 3.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.5 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.2 | 1.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 1.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 1.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.7 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0016418 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 0.1 | 1.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 1.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 6.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 1.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 10.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 4.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 3.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 1.8 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 1.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 10.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 7.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.9 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 1.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 2.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 2.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 4.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 4.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 7.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 8.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 8.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 8.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 3.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 3.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 6.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 4.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 6.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 1.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 6.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.7 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |