Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tead1
Z-value: 1.32
Transcription factors associated with Tead1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead1
|
ENSMUSG00000055320.19 | Tead1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead1 | mm39_v1_chr7_+_112341233_112341268 | 0.70 | 1.2e-11 | Click! |
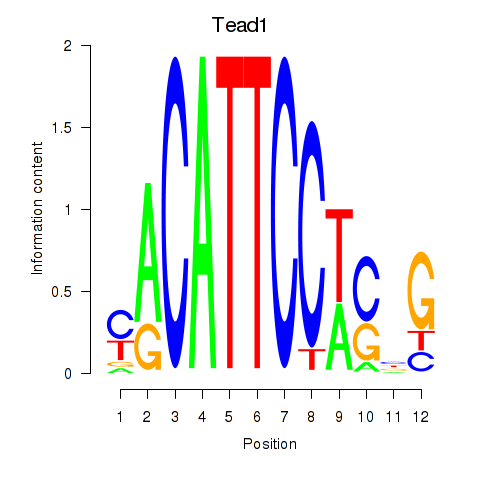
Activity profile of Tead1 motif
Sorted Z-values of Tead1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_17306334 | 11.04 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr6_+_17307038 | 9.59 |
ENSMUST00000123439.8
|
Cav1
|
caveolin 1, caveolae protein |
| chr1_+_135764092 | 9.48 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr6_+_17306379 | 9.38 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr17_+_35960600 | 9.36 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr9_-_107486381 | 8.41 |
ENSMUST00000102531.7
ENSMUST00000102530.8 ENSMUST00000195057.2 ENSMUST00000102532.10 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr6_+_17306414 | 8.32 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr11_-_100861713 | 8.23 |
ENSMUST00000060792.6
|
Cavin1
|
caveolae associated 1 |
| chr14_-_55204054 | 7.82 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr14_-_55204092 | 7.78 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr15_+_78726824 | 7.09 |
ENSMUST00000059619.3
|
Cdc42ep1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr19_-_23425757 | 6.56 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr2_+_156617329 | 6.41 |
ENSMUST00000088552.7
|
Myl9
|
myosin, light polypeptide 9, regulatory |
| chr14_-_55204383 | 6.35 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr11_+_62711057 | 6.27 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr1_+_192984278 | 6.23 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr1_+_189460461 | 5.81 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr6_+_113435716 | 5.41 |
ENSMUST00000203661.3
ENSMUST00000204774.3 ENSMUST00000053569.7 ENSMUST00000101065.8 |
Il17re
|
interleukin 17 receptor E |
| chr11_+_62711295 | 5.30 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr7_-_33832640 | 5.23 |
ENSMUST00000038537.9
|
Wtip
|
WT1-interacting protein |
| chr7_+_140796537 | 5.16 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr14_-_55204023 | 5.12 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr17_-_71309815 | 4.98 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr8_+_129450766 | 4.98 |
ENSMUST00000149116.2
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta) |
| chr19_-_57185928 | 4.95 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr14_-_55231998 | 4.83 |
ENSMUST00000227518.2
ENSMUST00000226424.2 ENSMUST00000153783.2 ENSMUST00000102803.11 ENSMUST00000168485.8 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr5_+_136996713 | 4.74 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr3_-_69767208 | 4.71 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr9_-_8004586 | 4.46 |
ENSMUST00000086580.12
ENSMUST00000065353.13 |
Yap1
|
yes-associated protein 1 |
| chr16_-_38620688 | 4.31 |
ENSMUST00000057767.6
|
Upk1b
|
uroplakin 1B |
| chr15_+_6416229 | 4.31 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr19_-_57185861 | 4.18 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr6_-_128277701 | 4.18 |
ENSMUST00000143004.2
ENSMUST00000006311.13 ENSMUST00000112157.4 ENSMUST00000133118.2 |
Tead4
|
TEA domain family member 4 |
| chr19_-_57185988 | 4.13 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr12_+_85793313 | 4.01 |
ENSMUST00000040461.4
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor 2 |
| chr6_+_146789978 | 3.96 |
ENSMUST00000016631.14
ENSMUST00000203730.3 ENSMUST00000111623.9 |
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr19_-_57185808 | 3.94 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr5_+_103573367 | 3.84 |
ENSMUST00000048957.11
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr14_-_54815000 | 3.76 |
ENSMUST00000054487.10
|
Ajuba
|
ajuba LIM protein |
| chr15_+_6416079 | 3.62 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr10_+_93476903 | 3.60 |
ENSMUST00000020204.5
|
Ntn4
|
netrin 4 |
| chr9_-_44624496 | 3.54 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr1_+_51328265 | 3.52 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr11_-_101061153 | 3.50 |
ENSMUST00000123864.2
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr10_+_24471340 | 3.46 |
ENSMUST00000020171.12
|
Ccn2
|
cellular communication network factor 2 |
| chr10_+_32959472 | 3.43 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr7_+_3340013 | 3.37 |
ENSMUST00000204541.2
|
Myadm
|
myeloid-associated differentiation marker |
| chr11_-_55310724 | 3.24 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr2_+_90948481 | 3.17 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_119173348 | 3.12 |
ENSMUST00000187474.7
ENSMUST00000187940.7 |
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr18_+_50186349 | 3.09 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_135693818 | 3.04 |
ENSMUST00000038945.6
|
Phlda3
|
pleckstrin homology like domain, family A, member 3 |
| chr6_+_125297596 | 3.04 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr7_+_130247912 | 2.87 |
ENSMUST00000207549.2
ENSMUST00000209108.2 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_87322443 | 2.85 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr6_+_34575435 | 2.83 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr6_-_37419030 | 2.81 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr17_-_71309012 | 2.74 |
ENSMUST00000128179.2
ENSMUST00000150456.2 ENSMUST00000233357.2 ENSMUST00000233417.2 |
Myl12a
Gm49909
|
myosin, light chain 12A, regulatory, non-sarcomeric predicted gene, 49909 |
| chr6_-_42350188 | 2.69 |
ENSMUST00000073387.5
ENSMUST00000204357.2 |
Epha1
|
Eph receptor A1 |
| chr7_+_43441315 | 2.69 |
ENSMUST00000005891.7
|
Klk9
|
kallikrein related-peptidase 9 |
| chr18_+_50164043 | 2.65 |
ENSMUST00000145726.2
ENSMUST00000128377.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_-_104324035 | 2.58 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr7_+_140796096 | 2.53 |
ENSMUST00000153081.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr10_-_12702674 | 2.48 |
ENSMUST00000219130.2
|
Utrn
|
utrophin |
| chr7_+_27196760 | 2.41 |
ENSMUST00000238964.2
|
Prx
|
periaxin |
| chr8_-_123278054 | 2.39 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr1_-_168259710 | 2.31 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr5_+_24631829 | 2.19 |
ENSMUST00000155598.8
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr2_-_60711706 | 2.17 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_+_90865958 | 2.14 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr6_-_128101275 | 2.13 |
ENSMUST00000127105.8
|
Tspan9
|
tetraspanin 9 |
| chr12_-_98703664 | 2.12 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr5_-_86616849 | 2.09 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr7_-_65020655 | 1.98 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr17_+_35878332 | 1.97 |
ENSMUST00000044326.5
|
2300002M23Rik
|
RIKEN cDNA 2300002M23 gene |
| chr4_+_154321982 | 1.96 |
ENSMUST00000152159.8
|
Megf6
|
multiple EGF-like-domains 6 |
| chr11_+_70591299 | 1.90 |
ENSMUST00000152618.9
ENSMUST00000102554.8 ENSMUST00000094499.11 ENSMUST00000072187.12 ENSMUST00000137119.3 |
Kif1c
|
kinesin family member 1C |
| chr10_+_27950809 | 1.71 |
ENSMUST00000166468.2
ENSMUST00000218359.2 ENSMUST00000218276.2 |
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr6_-_115569504 | 1.70 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr11_+_115865535 | 1.69 |
ENSMUST00000021107.14
ENSMUST00000169928.8 ENSMUST00000106461.8 |
Itgb4
|
integrin beta 4 |
| chr7_-_109215754 | 1.57 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr8_+_86567600 | 1.56 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr3_+_96011810 | 1.54 |
ENSMUST00000132980.8
ENSMUST00000138206.8 ENSMUST00000090785.9 ENSMUST00000035519.12 |
Otud7b
|
OTU domain containing 7B |
| chr5_+_31274046 | 1.51 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr4_+_62278932 | 1.49 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr7_-_143056252 | 1.46 |
ENSMUST00000010904.5
|
Phlda2
|
pleckstrin homology like domain, family A, member 2 |
| chr11_+_77353218 | 1.43 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr7_-_28071919 | 1.39 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr5_+_31274064 | 1.36 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr11_+_68961599 | 1.32 |
ENSMUST00000075980.12
ENSMUST00000094081.5 |
Tmem107
|
transmembrane protein 107 |
| chr12_+_55349422 | 1.28 |
ENSMUST00000021411.15
|
Prorp
|
protein only RNase P catalytic subunit |
| chr3_+_31049896 | 1.27 |
ENSMUST00000108249.9
|
Prkci
|
protein kinase C, iota |
| chr17_-_48145466 | 1.19 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr12_+_9023892 | 1.12 |
ENSMUST00000085745.13
ENSMUST00000111113.3 |
Wdr35
|
WD repeat domain 35 |
| chr8_+_122985359 | 1.03 |
ENSMUST00000187142.3
|
Zfp469
|
zinc finger protein 469 |
| chr7_-_28071658 | 1.01 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_+_46081213 | 0.87 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_102875229 | 0.84 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr9_-_65298934 | 0.83 |
ENSMUST00000068307.4
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr8_-_96033213 | 0.81 |
ENSMUST00000119870.9
ENSMUST00000093268.5 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr4_+_149629559 | 0.79 |
ENSMUST00000105692.2
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr11_-_83320281 | 0.76 |
ENSMUST00000052521.9
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr5_+_77099229 | 0.71 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr5_-_21850579 | 0.71 |
ENSMUST00000051358.11
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr1_+_156138286 | 0.69 |
ENSMUST00000027896.10
|
Nphs2
|
nephrosis 2, podocin |
| chr2_+_20524587 | 0.68 |
ENSMUST00000114604.9
ENSMUST00000066509.10 |
Etl4
|
enhancer trap locus 4 |
| chrX_-_6232775 | 0.64 |
ENSMUST00000024049.8
|
Bmp15
|
bone morphogenetic protein 15 |
| chr1_+_4878460 | 0.64 |
ENSMUST00000131119.2
|
Lypla1
|
lysophospholipase 1 |
| chr5_-_115438971 | 0.64 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr10_-_42152684 | 0.63 |
ENSMUST00000175881.8
ENSMUST00000056974.4 ENSMUST00000105502.8 ENSMUST00000105501.2 |
Foxo3
|
forkhead box O3 |
| chr8_+_70755402 | 0.61 |
ENSMUST00000150968.8
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr17_-_71575584 | 0.57 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr8_-_105036664 | 0.52 |
ENSMUST00000160596.8
ENSMUST00000164175.2 |
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr4_-_62278761 | 0.47 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr19_-_42117420 | 0.46 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chr7_+_140796559 | 0.41 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr4_+_43267165 | 0.40 |
ENSMUST00000107942.9
ENSMUST00000102953.4 |
Atp8b5
|
ATPase, class I, type 8B, member 5 |
| chr18_+_62086122 | 0.39 |
ENSMUST00000051720.6
ENSMUST00000235860.2 |
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr11_+_115865756 | 0.36 |
ENSMUST00000106460.9
|
Itgb4
|
integrin beta 4 |
| chr8_-_106692668 | 0.34 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr5_-_30401416 | 0.32 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr8_+_70755168 | 0.31 |
ENSMUST00000066469.14
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr8_-_48443525 | 0.31 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr18_-_9449947 | 0.31 |
ENSMUST00000234102.2
|
Ccny
|
cyclin Y |
| chr8_-_105036739 | 0.30 |
ENSMUST00000159039.2
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr6_-_120341304 | 0.28 |
ENSMUST00000146667.2
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr17_-_13271183 | 0.28 |
ENSMUST00000091648.4
|
Gpr31b
|
G protein-coupled receptor 31, D17Leh66b region |
| chr6_-_134543876 | 0.28 |
ENSMUST00000032322.15
ENSMUST00000126836.4 |
Lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr8_-_70755132 | 0.19 |
ENSMUST00000008004.10
|
Ddx49
|
DEAD box helicase 49 |
| chr14_-_123151162 | 0.17 |
ENSMUST00000160401.8
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr18_-_35795175 | 0.17 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr2_-_32665596 | 0.15 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr18_-_9450097 | 0.14 |
ENSMUST00000053917.6
|
Ccny
|
cyclin Y |
| chr4_+_58285957 | 0.12 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr4_-_139695337 | 0.11 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr5_-_135518098 | 0.10 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr4_-_62278673 | 0.09 |
ENSMUST00000084527.10
ENSMUST00000098033.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr2_-_120234539 | 0.09 |
ENSMUST00000090042.12
ENSMUST00000110729.2 ENSMUST00000090046.12 |
Tmem87a
|
transmembrane protein 87A |
| chr8_+_84267722 | 0.09 |
ENSMUST00000058609.5
|
Olfr370
|
olfactory receptor 370 |
| chr7_+_43645253 | 0.03 |
ENSMUST00000007156.5
|
Klk1b11
|
kallikrein 1-related peptidase b11 |
| chr7_-_44174065 | 0.03 |
ENSMUST00000165208.4
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr7_+_98092628 | 0.01 |
ENSMUST00000098274.5
|
Gucy2d
|
guanylate cyclase 2d |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.1 | GO:0007522 | visceral muscle development(GO:0007522) |
| 6.4 | 38.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 2.4 | 9.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 2.1 | 8.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 1.7 | 5.0 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 1.6 | 4.8 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 1.3 | 7.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.9 | 9.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.9 | 11.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.8 | 4.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.6 | 4.7 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.6 | 3.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.5 | 6.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 4.5 | GO:0060449 | contact inhibition(GO:0060242) bud elongation involved in lung branching(GO:0060449) |
| 0.5 | 1.5 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.5 | 2.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 3.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 7.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.4 | 2.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.4 | 2.5 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 2.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 2.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.3 | 1.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 1.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 3.5 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 1.5 | GO:0072719 | cellular response to cisplatin(GO:0072719) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.2 | 5.8 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.2 | 3.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.1 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 17.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 2.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 8.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.9 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 1.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 3.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.3 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.6 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.1 | 3.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 6.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 2.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 5.7 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.1 | 3.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 2.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 2.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 3.0 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 1.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 3.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 2.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 8.1 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 2.8 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 3.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 3.8 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 3.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 1.6 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 5.0 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.7 | GO:0007588 | excretion(GO:0007588) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 38.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.7 | 5.0 | GO:0034681 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha7-beta1 complex(GO:0034677) integrin alpha10-beta1 complex(GO:0034680) integrin alpha11-beta1 complex(GO:0034681) |
| 1.6 | 7.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.6 | 14.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 1.4 | 9.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.3 | 35.1 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 1.1 | 4.5 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 1.0 | 6.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.9 | 3.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.8 | 3.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 2.2 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.5 | 4.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.3 | 2.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 8.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 3.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 16.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 14.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 3.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 2.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 11.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 8.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 3.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 3.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 6.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 38.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 3.9 | 27.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 1.6 | 9.5 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.2 | 8.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.9 | 4.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.9 | 11.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.8 | 5.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 4.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.6 | 5.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 2.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 9.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 3.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 8.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 7.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 2.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 2.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 7.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 3.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 3.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 4.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 2.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 3.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 3.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 4.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 5.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 7.7 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 3.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 5.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 3.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 15.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 4.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 4.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 3.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 38.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 8.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 12.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 6.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 4.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 35.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 36.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 17.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.4 | 5.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 4.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 8.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 6.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 8.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 3.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |