Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tead3_Tead4
Z-value: 3.77
Transcription factors associated with Tead3_Tead4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
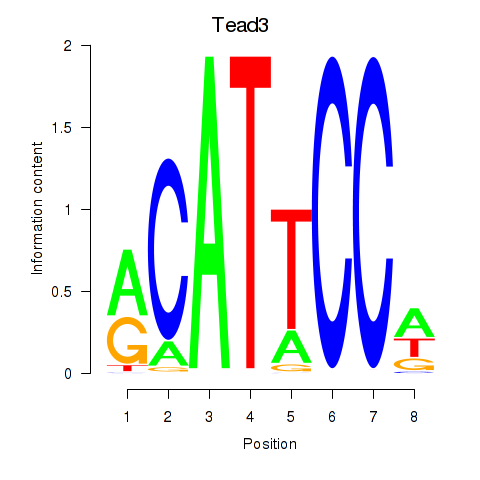
Tead3
|
ENSMUSG00000002249.22 | Tead3 |
|
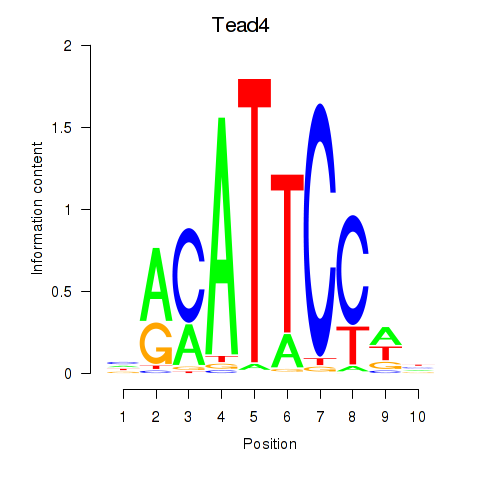
Tead4
|
ENSMUSG00000030353.16 | Tead4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead4 | mm39_v1_chr6_-_128277701_128277786 | 0.77 | 2.0e-15 | Click! |
| Tead3 | mm39_v1_chr17_-_28569574_28569685 | 0.70 | 9.7e-12 | Click! |
Activity profile of Tead3_Tead4 motif
Sorted Z-values of Tead3_Tead4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead3_Tead4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_38182569 | 54.50 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr10_+_24471340 | 43.94 |
ENSMUST00000020171.12
|
Ccn2
|
cellular communication network factor 2 |
| chr9_-_66956425 | 34.87 |
ENSMUST00000113687.8
ENSMUST00000113693.8 ENSMUST00000113701.8 ENSMUST00000034928.12 ENSMUST00000113685.10 ENSMUST00000030185.5 ENSMUST00000050905.16 ENSMUST00000113705.8 ENSMUST00000113697.8 ENSMUST00000113707.9 |
Tpm1
|
tropomyosin 1, alpha |
| chr10_+_53213763 | 29.94 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr18_+_23548534 | 27.59 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr15_+_6416229 | 26.04 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr11_-_100861713 | 25.53 |
ENSMUST00000060792.6
|
Cavin1
|
caveolae associated 1 |
| chr8_+_123912976 | 23.41 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr8_-_62213100 | 21.59 |
ENSMUST00000121493.9
|
Palld
|
palladin, cytoskeletal associated protein |
| chr2_+_25346841 | 21.36 |
ENSMUST00000114265.9
ENSMUST00000102918.3 |
Clic3
|
chloride intracellular channel 3 |
| chr14_-_55231998 | 21.23 |
ENSMUST00000227518.2
ENSMUST00000226424.2 ENSMUST00000153783.2 ENSMUST00000102803.11 ENSMUST00000168485.8 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr6_-_67512768 | 20.30 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr1_+_135764092 | 20.13 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr17_+_71326510 | 19.89 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr1_+_51328265 | 19.14 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr7_-_33832640 | 19.08 |
ENSMUST00000038537.9
|
Wtip
|
WT1-interacting protein |
| chr1_+_189460461 | 18.92 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr6_-_128277701 | 18.90 |
ENSMUST00000143004.2
ENSMUST00000006311.13 ENSMUST00000112157.4 ENSMUST00000133118.2 |
Tead4
|
TEA domain family member 4 |
| chr6_+_17307038 | 18.68 |
ENSMUST00000123439.8
|
Cav1
|
caveolin 1, caveolae protein |
| chr17_+_71326542 | 18.25 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr18_+_23548455 | 17.42 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr9_-_107482494 | 17.09 |
ENSMUST00000102529.10
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr17_+_35960600 | 16.81 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr14_-_55204092 | 16.62 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr14_-_55204054 | 16.52 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr6_+_17306334 | 16.09 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr17_-_71309815 | 16.01 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_+_98303287 | 15.99 |
ENSMUST00000058295.6
|
Erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr13_-_113182891 | 15.87 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr9_-_107486381 | 15.59 |
ENSMUST00000102531.7
ENSMUST00000102530.8 ENSMUST00000195057.2 ENSMUST00000102532.10 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr14_-_54815000 | 14.91 |
ENSMUST00000054487.10
|
Ajuba
|
ajuba LIM protein |
| chr1_-_134731516 | 14.49 |
ENSMUST00000238280.2
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_22696025 | 14.42 |
ENSMUST00000218002.2
ENSMUST00000049930.9 |
Tcf21
|
transcription factor 21 |
| chr6_+_17306379 | 14.38 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr7_-_44174065 | 14.38 |
ENSMUST00000165208.4
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr3_-_151971391 | 14.24 |
ENSMUST00000199470.5
ENSMUST00000200589.5 |
Nexn
|
nexilin |
| chr15_+_6416079 | 14.07 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr5_+_31274046 | 14.00 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_31274064 | 13.84 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr6_+_113435716 | 13.56 |
ENSMUST00000203661.3
ENSMUST00000204774.3 ENSMUST00000053569.7 ENSMUST00000101065.8 |
Il17re
|
interleukin 17 receptor E |
| chr1_+_45350698 | 13.51 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr1_-_72914036 | 13.38 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr14_-_55204383 | 13.21 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr4_+_148070245 | 13.20 |
ENSMUST00000103231.5
|
Nppb
|
natriuretic peptide type B |
| chr13_+_49761506 | 13.10 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr6_+_122490534 | 12.79 |
ENSMUST00000032210.14
ENSMUST00000148517.8 |
Mfap5
|
microfibrillar associated protein 5 |
| chr6_+_17306414 | 12.54 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr9_-_107482462 | 12.33 |
ENSMUST00000194433.6
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr10_-_78427721 | 12.26 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr5_-_137312424 | 12.19 |
ENSMUST00000199121.2
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr10_+_93476903 | 12.08 |
ENSMUST00000020204.5
|
Ntn4
|
netrin 4 |
| chr6_+_29433247 | 12.08 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr6_+_146789978 | 12.04 |
ENSMUST00000016631.14
ENSMUST00000203730.3 ENSMUST00000111623.9 |
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr1_+_170983081 | 11.95 |
ENSMUST00000111334.2
|
Mpz
|
myelin protein zero |
| chr5_+_67125759 | 11.91 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr6_+_122490635 | 11.85 |
ENSMUST00000142896.8
ENSMUST00000121656.2 |
Mfap5
|
microfibrillar associated protein 5 |
| chr17_+_47747657 | 11.76 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr18_-_35855383 | 11.63 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr17_-_28569574 | 11.47 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr17_+_47747540 | 11.39 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr6_+_122490577 | 11.18 |
ENSMUST00000118626.8
|
Mfap5
|
microfibrillar associated protein 5 |
| chr2_+_90948481 | 11.15 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr8_-_123278054 | 10.95 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr2_+_129854256 | 10.68 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr5_-_18054702 | 10.68 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr15_+_78726824 | 10.64 |
ENSMUST00000059619.3
|
Cdc42ep1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr12_+_75355082 | 10.59 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr11_-_101061153 | 10.52 |
ENSMUST00000123864.2
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr6_-_119173348 | 10.51 |
ENSMUST00000187474.7
ENSMUST00000187940.7 |
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr14_-_55204023 | 10.46 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr4_+_94627755 | 10.40 |
ENSMUST00000071168.6
|
Tek
|
TEK receptor tyrosine kinase |
| chr5_-_18054781 | 10.38 |
ENSMUST00000170051.8
|
Cd36
|
CD36 molecule |
| chr2_+_74522258 | 9.94 |
ENSMUST00000061745.5
|
Hoxd10
|
homeobox D10 |
| chr19_-_23425757 | 9.88 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chrX_+_132809166 | 9.80 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr14_+_30601157 | 9.78 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr8_-_124621483 | 9.73 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr12_-_98703664 | 9.71 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr4_+_94627513 | 9.70 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr19_+_53781721 | 9.63 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr15_-_12549350 | 9.62 |
ENSMUST00000190929.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr11_-_32172233 | 9.59 |
ENSMUST00000150381.2
ENSMUST00000144902.2 ENSMUST00000020524.15 |
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr10_-_63039709 | 9.58 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chr3_-_145355725 | 9.45 |
ENSMUST00000029846.5
|
Ccn1
|
cellular communication network factor 1 |
| chr4_+_134042423 | 9.37 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr19_-_36097233 | 9.23 |
ENSMUST00000025718.10
|
Ankrd1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr11_-_115704447 | 9.20 |
ENSMUST00000041684.11
ENSMUST00000156812.2 |
Caskin2
|
CASK-interacting protein 2 |
| chr1_-_74040723 | 9.17 |
ENSMUST00000190389.7
|
Tns1
|
tensin 1 |
| chr3_-_129548954 | 9.10 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr5_+_104607316 | 9.09 |
ENSMUST00000086831.4
|
Pkd2
|
polycystin 2, transient receptor potential cation channel |
| chr5_-_69699965 | 9.06 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr2_-_148285450 | 9.04 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr1_+_135252508 | 9.04 |
ENSMUST00000059352.3
|
Lmod1
|
leiomodin 1 (smooth muscle) |
| chr11_+_70591299 | 9.00 |
ENSMUST00000152618.9
ENSMUST00000102554.8 ENSMUST00000094499.11 ENSMUST00000072187.12 ENSMUST00000137119.3 |
Kif1c
|
kinesin family member 1C |
| chr2_-_181313415 | 8.89 |
ENSMUST00000054491.6
|
Sox18
|
SRY (sex determining region Y)-box 18 |
| chr19_+_8966641 | 8.82 |
ENSMUST00000092956.4
ENSMUST00000092955.11 |
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr8_-_48443525 | 8.75 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr11_+_77353218 | 8.72 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr3_-_57483175 | 8.68 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chrX_+_132809189 | 8.66 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr3_-_92734546 | 8.59 |
ENSMUST00000072363.5
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr11_+_96214078 | 8.53 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr5_-_103777145 | 8.40 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr9_-_58065800 | 8.36 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr8_-_86091970 | 8.33 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr7_-_65020655 | 8.14 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr3_-_57483330 | 8.14 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr2_+_72115981 | 8.04 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr10_+_32959472 | 7.99 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr10_+_5543769 | 7.95 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr7_+_43441315 | 7.79 |
ENSMUST00000005891.7
|
Klk9
|
kallikrein related-peptidase 9 |
| chr17_-_28569721 | 7.72 |
ENSMUST00000156862.3
|
Tead3
|
TEA domain family member 3 |
| chr13_-_63006176 | 7.69 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr15_+_101308935 | 7.63 |
ENSMUST00000147662.8
|
Krt7
|
keratin 7 |
| chr7_-_30754223 | 7.55 |
ENSMUST00000206012.2
ENSMUST00000108110.5 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr14_-_32407203 | 7.52 |
ENSMUST00000096038.4
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr5_-_69699932 | 7.48 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr7_+_128125339 | 7.44 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr2_-_60383647 | 7.43 |
ENSMUST00000112525.5
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr2_-_17465410 | 7.42 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr6_-_37419030 | 7.38 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr1_-_153061758 | 7.34 |
ENSMUST00000185356.7
|
Lamc2
|
laminin, gamma 2 |
| chr7_-_30754193 | 7.34 |
ENSMUST00000205778.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr3_+_19698631 | 7.32 |
ENSMUST00000029139.9
|
Trim55
|
tripartite motif-containing 55 |
| chr5_-_24806960 | 7.26 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr5_+_53206688 | 7.24 |
ENSMUST00000094787.8
|
Slc34a2
|
solute carrier family 34 (sodium phosphate), member 2 |
| chr11_-_5753693 | 7.24 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr17_-_24863907 | 7.21 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr11_+_96740689 | 7.21 |
ENSMUST00000018816.14
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chr2_+_90865958 | 7.20 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr8_-_86091946 | 7.20 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr19_-_57185928 | 7.16 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr9_+_102594867 | 7.14 |
ENSMUST00000035121.14
|
Amotl2
|
angiomotin-like 2 |
| chr2_+_160722562 | 7.11 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chr7_-_30754240 | 7.08 |
ENSMUST00000206860.2
ENSMUST00000071697.11 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr18_+_50164043 | 7.05 |
ENSMUST00000145726.2
ENSMUST00000128377.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_-_57983511 | 7.04 |
ENSMUST00000173990.8
ENSMUST00000022531.14 |
Lats2
|
large tumor suppressor 2 |
| chr1_+_43769750 | 6.97 |
ENSMUST00000027217.9
|
Ecrg4
|
ECRG4 augurin precursor |
| chr16_-_95387444 | 6.93 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor |
| chr17_+_88933957 | 6.92 |
ENSMUST00000163588.8
ENSMUST00000064035.13 |
Ston1
|
stonin 1 |
| chr9_+_69361348 | 6.87 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr15_-_12549963 | 6.87 |
ENSMUST00000189324.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr13_+_112600604 | 6.80 |
ENSMUST00000183663.8
ENSMUST00000184311.8 ENSMUST00000183886.8 |
Il6st
|
interleukin 6 signal transducer |
| chr12_+_85793313 | 6.76 |
ENSMUST00000040461.4
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor 2 |
| chr7_+_130247912 | 6.75 |
ENSMUST00000207549.2
ENSMUST00000209108.2 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_-_105350898 | 6.74 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr16_-_38620688 | 6.70 |
ENSMUST00000057767.6
|
Upk1b
|
uroplakin 1B |
| chr6_-_142453531 | 6.67 |
ENSMUST00000134191.3
ENSMUST00000239397.2 ENSMUST00000239395.2 ENSMUST00000032373.12 |
Ldhb
|
lactate dehydrogenase B |
| chr8_+_94863825 | 6.60 |
ENSMUST00000034207.8
|
Mt4
|
metallothionein 4 |
| chr3_-_116601451 | 6.59 |
ENSMUST00000159670.3
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr18_+_23548192 | 6.58 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr6_-_52217821 | 6.57 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr6_+_145879839 | 6.51 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr1_+_135693818 | 6.47 |
ENSMUST00000038945.6
|
Phlda3
|
pleckstrin homology like domain, family A, member 3 |
| chr5_+_67055753 | 6.42 |
ENSMUST00000122812.5
ENSMUST00000117601.8 |
Limch1
|
LIM and calponin homology domains 1 |
| chr9_-_8004586 | 6.41 |
ENSMUST00000086580.12
ENSMUST00000065353.13 |
Yap1
|
yes-associated protein 1 |
| chr13_-_95661726 | 6.37 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr11_+_100306523 | 6.34 |
ENSMUST00000001595.10
ENSMUST00000107400.3 |
Fkbp10
|
FK506 binding protein 10 |
| chr6_+_34575435 | 6.29 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr14_-_45715308 | 6.28 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr15_-_42540363 | 6.28 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr4_+_154321982 | 6.25 |
ENSMUST00000152159.8
|
Megf6
|
multiple EGF-like-domains 6 |
| chr19_-_57185861 | 6.25 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr1_-_66984178 | 6.23 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_+_75362187 | 6.23 |
ENSMUST00000137868.8
|
Speg
|
SPEG complex locus |
| chr3_-_101017594 | 6.20 |
ENSMUST00000102694.4
|
Ptgfrn
|
prostaglandin F2 receptor negative regulator |
| chr17_+_88933997 | 6.19 |
ENSMUST00000150023.2
|
Ston1
|
stonin 1 |
| chrX_+_72030945 | 6.15 |
ENSMUST00000164800.8
ENSMUST00000114546.9 |
Zfp185
|
zinc finger protein 185 |
| chr14_+_101967343 | 6.12 |
ENSMUST00000100337.10
|
Lmo7
|
LIM domain only 7 |
| chr9_-_58943670 | 6.10 |
ENSMUST00000214547.2
ENSMUST00000068664.7 |
Neo1
|
neogenin |
| chr4_+_118286898 | 6.08 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr8_-_95644639 | 6.01 |
ENSMUST00000077955.6
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr1_-_66984521 | 6.00 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr3_-_116601700 | 5.99 |
ENSMUST00000159742.8
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr10_+_21868114 | 5.96 |
ENSMUST00000150089.8
ENSMUST00000100036.10 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_-_30754792 | 5.88 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr4_-_58206596 | 5.85 |
ENSMUST00000042850.9
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr9_-_18585826 | 5.84 |
ENSMUST00000208663.2
|
Muc16
|
mucin 16 |
| chr3_-_154760978 | 5.82 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr3_-_103645311 | 5.78 |
ENSMUST00000029440.10
|
Olfml3
|
olfactomedin-like 3 |
| chr19_-_57185988 | 5.78 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr12_+_119354110 | 5.75 |
ENSMUST00000222058.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr4_+_97665992 | 5.73 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr7_+_144854542 | 5.72 |
ENSMUST00000033386.12
|
Mrgprf
|
MAS-related GPR, member F |
| chr17_-_71309012 | 5.71 |
ENSMUST00000128179.2
ENSMUST00000150456.2 ENSMUST00000233357.2 ENSMUST00000233417.2 |
Myl12a
Gm49909
|
myosin, light chain 12A, regulatory, non-sarcomeric predicted gene, 49909 |
| chr9_+_102594474 | 5.67 |
ENSMUST00000153911.2
|
Amotl2
|
angiomotin-like 2 |
| chr19_-_57185808 | 5.66 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr9_-_44624496 | 5.61 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr17_+_75772475 | 5.60 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr1_+_12762501 | 5.53 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr19_+_45006552 | 5.40 |
ENSMUST00000237043.2
ENSMUST00000178087.3 |
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr9_-_77255099 | 5.31 |
ENSMUST00000184138.8
ENSMUST00000184006.8 ENSMUST00000185144.8 ENSMUST00000034910.16 |
Mlip
|
muscular LMNA-interacting protein |
| chr2_+_155118217 | 5.27 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr4_+_57637817 | 5.22 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr7_+_141048722 | 5.22 |
ENSMUST00000058746.7
|
Cd151
|
CD151 antigen |
| chr14_+_47120311 | 5.18 |
ENSMUST00000022386.15
ENSMUST00000228404.2 ENSMUST00000100672.11 |
Samd4
|
sterile alpha motif domain containing 4 |
| chr5_-_135518098 | 5.16 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr8_-_105350881 | 5.14 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr8_-_37081091 | 5.12 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr1_+_40619215 | 5.12 |
ENSMUST00000027233.9
|
Slc9a4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.2 | 56.8 | GO:0007522 | visceral muscle development(GO:0007522) |
| 10.9 | 54.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 10.3 | 61.7 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 7.1 | 21.2 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 6.7 | 40.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 6.4 | 25.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 5.9 | 23.4 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 5.8 | 34.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 5.3 | 26.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 5.0 | 20.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 4.8 | 14.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 4.5 | 44.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 4.2 | 21.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 4.1 | 12.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 3.9 | 15.5 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 3.8 | 19.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 3.8 | 18.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 3.7 | 63.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 3.3 | 13.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 3.2 | 9.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 3.2 | 16.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) estrous cycle(GO:0044849) |
| 3.1 | 9.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 3.0 | 9.1 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 3.0 | 29.9 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 2.9 | 8.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 2.7 | 13.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.7 | 8.1 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 2.5 | 27.9 | GO:1903797 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 2.5 | 12.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.3 | 7.0 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 2.3 | 23.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 2.3 | 6.8 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 2.2 | 10.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.2 | 23.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.1 | 6.4 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 2.0 | 8.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 1.9 | 11.7 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 1.9 | 7.7 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.8 | 8.9 | GO:0048866 | stem cell fate specification(GO:0048866) |
| 1.7 | 8.4 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 1.6 | 19.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 1.5 | 4.6 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.5 | 4.6 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 1.5 | 12.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.5 | 7.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.4 | 7.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.4 | 4.3 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 1.4 | 8.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.4 | 21.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 1.3 | 8.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 1.3 | 35.8 | GO:0097435 | fibril organization(GO:0097435) |
| 1.3 | 18.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 1.3 | 2.6 | GO:0072199 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.3 | 9.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 1.3 | 5.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 1.3 | 1.3 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 1.2 | 9.8 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 1.2 | 7.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.2 | 8.3 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 1.2 | 6.9 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 1.2 | 4.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 1.1 | 4.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.1 | 3.4 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.1 | 5.5 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 1.1 | 4.3 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 1.1 | 4.3 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 1.0 | 7.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.0 | 2.9 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 1.0 | 10.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.0 | 4.8 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.9 | 21.6 | GO:0003334 | keratinocyte development(GO:0003334) positive regulation of podosome assembly(GO:0071803) |
| 0.9 | 4.4 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.9 | 3.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.9 | 2.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.9 | 18.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.8 | 9.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 7.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.8 | 11.9 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.8 | 9.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.8 | 7.9 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.8 | 1.5 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.7 | 4.5 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.7 | 4.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.7 | 2.2 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.7 | 13.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.7 | 2.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.7 | 15.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.7 | 17.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.6 | 5.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.6 | 2.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.6 | 17.2 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.6 | 6.7 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.6 | 4.7 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.6 | 1.7 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.6 | 5.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.5 | 3.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.5 | 9.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.5 | 2.1 | GO:0010735 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.5 | 13.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.5 | 11.6 | GO:0030953 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.5 | 9.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.5 | 6.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 6.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 17.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 | 4.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 | 18.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.5 | 1.4 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.5 | 0.9 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.5 | 1.4 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.5 | 8.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.4 | 1.7 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.4 | 4.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 | 2.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.4 | 5.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 1.7 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.4 | 4.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 15.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.4 | 4.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 7.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.4 | 6.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.4 | 1.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.4 | 21.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.3 | 7.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 3.3 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) intestinal epithelial cell development(GO:0060576) |
| 0.3 | 5.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.0 | GO:1904630 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.3 | 1.9 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.3 | 1.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 5.8 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.3 | 1.8 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.3 | 2.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 5.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.3 | 2.0 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.3 | 3.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 1.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.3 | 3.8 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.3 | 29.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.3 | 1.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 12.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 1.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 1.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 1.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 7.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 6.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.2 | 1.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.2 | 1.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 6.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 7.2 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 1.6 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 2.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 4.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 6.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 2.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 8.9 | GO:0014741 | negative regulation of muscle hypertrophy(GO:0014741) |
| 0.2 | 6.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 1.2 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 1.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 3.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 7.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 6.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.4 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.2 | 10.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.2 | 6.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 11.4 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.2 | 1.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 5.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 9.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.2 | 0.9 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.2 | 9.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.2 | 1.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 1.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 6.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 1.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 8.3 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.5 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.5 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 0.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 26.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 2.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 7.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 3.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 2.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 6.5 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 5.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 3.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.7 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.6 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.0 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 2.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.7 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 6.3 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 8.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 5.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.6 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 2.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 5.9 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 0.8 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 8.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.8 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 2.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 11.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 3.9 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 1.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 3.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.7 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.8 | GO:0051384 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.1 | 3.1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.4 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 3.1 | GO:0042107 | cytokine metabolic process(GO:0042107) |
| 0.0 | 2.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.2 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 1.4 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 4.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 3.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 3.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.4 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 1.2 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 1.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.2 | 51.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 8.0 | 40.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 5.2 | 141.7 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 3.9 | 11.7 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 3.6 | 60.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 2.9 | 20.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.8 | 25.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 2.8 | 8.3 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 2.4 | 7.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 2.3 | 34.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 2.3 | 6.8 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 2.1 | 35.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 2.0 | 8.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.6 | 6.4 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 1.5 | 4.6 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha7-beta1 complex(GO:0034677) integrin alpha10-beta1 complex(GO:0034680) integrin alpha11-beta1 complex(GO:0034681) |
| 1.5 | 6.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 1.4 | 8.6 | GO:0031523 | Myb complex(GO:0031523) |
| 1.3 | 13.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.3 | 9.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 1.2 | 34.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.1 | 16.0 | GO:0043219 | lateral loop(GO:0043219) |
| 1.0 | 13.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.0 | 5.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.7 | 91.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.7 | 7.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.6 | 6.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.6 | 96.1 | GO:0005901 | caveola(GO:0005901) |
| 0.6 | 12.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 4.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.5 | 7.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.5 | 12.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 3.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.5 | 10.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 2.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 8.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 7.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 12.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 52.3 | GO:0031674 | I band(GO:0031674) |
| 0.4 | 17.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 6.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.4 | 3.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.4 | 78.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.4 | 20.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 2.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 7.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 5.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 42.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.3 | 15.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 12.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 4.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 16.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 7.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 16.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 7.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 5.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 2.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 5.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 19.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 10.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.2 | 0.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 18.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 4.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 4.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 6.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 26.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 30.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 46.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 20.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 2.1 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 10.2 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.1 | 1.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 6.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 9.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 22.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 3.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 15.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 11.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 26.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0070161 | anchoring junction(GO:0070161) |
| 0.0 | 3.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0005715 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.0 | 6.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 5.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 4.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 6.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 24.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.0 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 61.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 9.0 | 35.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 8.1 | 56.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 5.7 | 17.2 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 5.3 | 16.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 4.3 | 25.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 4.3 | 29.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 4.3 | 72.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 4.2 | 21.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.6 | 25.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.3 | 23.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 2.4 | 7.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.4 | 45.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 2.3 | 6.8 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 2.1 | 40.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 1.8 | 9.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 1.7 | 15.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.6 | 6.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.5 | 12.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.5 | 13.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.4 | 7.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.4 | 8.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.4 | 13.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.4 | 9.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.3 | 9.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.3 | 11.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.2 | 8.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.2 | 9.4 | GO:0031432 | titin binding(GO:0031432) |
| 1.2 | 55.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.2 | 8.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.1 | 9.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 6.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.1 | 52.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 1.0 | 5.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.0 | 10.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 1.0 | 21.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.9 | 4.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.9 | 14.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.9 | 10.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.8 | 10.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.8 | 13.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.8 | 5.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.8 | 18.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.8 | 4.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 6.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.7 | 9.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 7.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 4.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.7 | 3.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.6 | 1.9 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.6 | 8.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 6.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.6 | 6.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 1.7 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 5.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 6.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 13.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.5 | 5.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 2.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.5 | 5.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 15.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 7.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 10.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 36.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.4 | 3.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 3.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.4 | 13.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.4 | 8.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 11.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 4.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 4.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.4 | 1.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.4 | 2.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 2.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.4 | 12.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 2.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 7.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 6.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 27.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 9.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 60.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 3.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 5.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 1.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.3 | 3.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 3.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 6.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 22.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.3 | 4.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 14.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.3 | 0.8 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 1.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 1.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 1.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 3.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 3.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 1.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 18.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 1.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 1.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 11.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 19.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 2.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.2 | 2.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 10.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 3.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 7.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 33.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 4.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 0.9 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.2 | 0.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 16.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 13.6 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 4.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 15.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 3.1 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 3.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 5.0 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 5.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 4.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 6.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 1.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 62.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 9.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 15.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 4.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 5.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 3.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 32.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 2.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 4.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 25.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 7.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 4.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 18.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 8.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 7.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 61.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.6 | 29.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.5 | 38.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.5 | 45.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.4 | 32.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.4 | 76.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 19.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 67.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 7.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 16.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 9.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 2.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 21.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 10.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 9.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 6.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 2.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 3.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 5.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 6.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 35.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 9.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 5.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 7.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 8.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 143.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 2.2 | 43.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 2.1 | 60.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.6 | 55.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.2 | 45.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.0 | 13.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.9 | 26.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.8 | 17.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.8 | 25.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.8 | 25.7 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.8 | 22.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.7 | 15.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 18.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.5 | 5.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 7.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 6.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 0.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.4 | 8.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.4 | 7.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 6.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 6.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 14.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 18.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 5.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 4.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 18.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 2.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 6.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.6 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 4.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 12.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 19.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 4.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 12.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |