Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
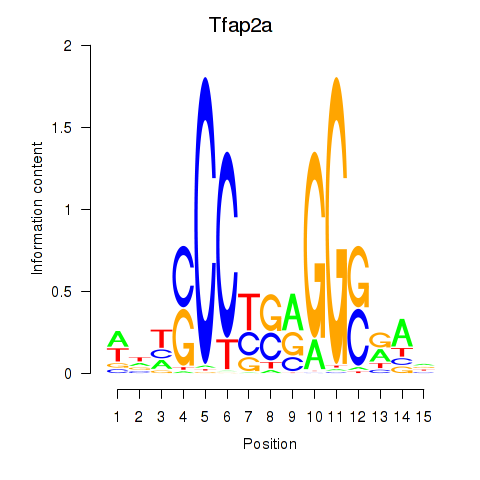
Results for Tfap2a
Z-value: 1.05
Transcription factors associated with Tfap2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2a
|
ENSMUSG00000021359.17 | Tfap2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2a | mm39_v1_chr13_-_40883893_40883919 | 0.32 | 5.7e-03 | Click! |
Activity profile of Tfap2a motif
Sorted Z-values of Tfap2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76869282 | 7.24 |
ENSMUST00000021459.14
|
Rab15
|
RAB15, member RAS oncogene family |
| chr11_-_103844870 | 5.68 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr15_-_44978223 | 5.56 |
ENSMUST00000022967.7
|
Kcnv1
|
potassium channel, subfamily V, member 1 |
| chr13_+_43276323 | 5.21 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_+_103254401 | 4.88 |
ENSMUST00000028609.14
|
Elf5
|
E74-like factor 5 |
| chr2_+_103254465 | 4.78 |
ENSMUST00000171693.8
|
Elf5
|
E74-like factor 5 |
| chr5_-_44139099 | 4.37 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chrX_+_10351360 | 4.37 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr5_-_44139121 | 4.31 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr2_+_119572770 | 4.30 |
ENSMUST00000028758.8
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr9_+_59614877 | 4.23 |
ENSMUST00000128944.8
ENSMUST00000098661.10 |
Gramd2
|
GRAM domain containing 2 |
| chr11_-_102787950 | 4.08 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr11_+_78213791 | 3.95 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr2_+_25179903 | 3.95 |
ENSMUST00000028337.7
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr13_-_110416637 | 3.93 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chr11_-_102787972 | 3.49 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr16_-_18445172 | 3.48 |
ENSMUST00000231335.2
ENSMUST00000232653.2 |
Gm49601
Septin5
|
predicted gene, 49601 septin 5 |
| chr9_+_109760856 | 3.43 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr7_+_97492124 | 3.40 |
ENSMUST00000033040.12
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr10_+_99851679 | 3.29 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr9_-_66951234 | 3.25 |
ENSMUST00000113690.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr7_-_30776081 | 3.19 |
ENSMUST00000072331.13
ENSMUST00000167369.8 |
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr14_-_65499835 | 3.15 |
ENSMUST00000131309.3
|
Fzd3
|
frizzled class receptor 3 |
| chr2_+_102380357 | 3.12 |
ENSMUST00000028612.8
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_+_83642910 | 3.09 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr13_+_25029088 | 3.04 |
ENSMUST00000006893.9
|
D130043K22Rik
|
RIKEN cDNA D130043K22 gene |
| chr10_-_76181089 | 3.01 |
ENSMUST00000036033.14
ENSMUST00000160048.8 ENSMUST00000105417.10 |
Dip2a
|
disco interacting protein 2 homolog A |
| chrX_-_8072714 | 3.01 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr5_+_107585774 | 3.00 |
ENSMUST00000162298.4
ENSMUST00000094541.4 ENSMUST00000211896.2 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr17_+_70276210 | 2.99 |
ENSMUST00000060072.12
|
Dlgap1
|
DLG associated protein 1 |
| chr7_+_30487322 | 2.98 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr1_+_191873078 | 2.95 |
ENSMUST00000078470.12
ENSMUST00000110844.3 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr8_+_126721878 | 2.91 |
ENSMUST00000046765.10
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr1_-_75254989 | 2.91 |
ENSMUST00000039534.11
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr4_-_155095441 | 2.89 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr8_+_126722113 | 2.87 |
ENSMUST00000212831.2
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr6_-_60805873 | 2.78 |
ENSMUST00000114268.5
|
Snca
|
synuclein, alpha |
| chr14_+_68321302 | 2.75 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr15_-_74599860 | 2.73 |
ENSMUST00000023261.4
ENSMUST00000190433.2 |
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr17_+_70276068 | 2.72 |
ENSMUST00000133983.8
|
Dlgap1
|
DLG associated protein 1 |
| chr5_-_52723700 | 2.71 |
ENSMUST00000039750.7
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chrX_-_102230225 | 2.71 |
ENSMUST00000121720.2
|
Nap1l2
|
nucleosome assembly protein 1-like 2 |
| chr6_+_36364990 | 2.70 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr11_-_118800314 | 2.67 |
ENSMUST00000117731.8
ENSMUST00000106278.9 ENSMUST00000120061.8 ENSMUST00000017576.11 |
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr15_-_37792237 | 2.65 |
ENSMUST00000168992.8
ENSMUST00000148652.9 |
Ncald
|
neurocalcin delta |
| chr7_-_3680530 | 2.64 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr18_+_61058684 | 2.64 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_+_43430459 | 2.63 |
ENSMUST00000014058.11
|
Klk10
|
kallikrein related-peptidase 10 |
| chr4_-_127224591 | 2.59 |
ENSMUST00000046532.4
|
Gjb3
|
gap junction protein, beta 3 |
| chrX_+_135723531 | 2.59 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr11_+_63022328 | 2.56 |
ENSMUST00000018361.10
|
Pmp22
|
peripheral myelin protein 22 |
| chr3_-_84387700 | 2.56 |
ENSMUST00000194027.2
ENSMUST00000107689.7 |
Fhdc1
|
FH2 domain containing 1 |
| chr9_-_31043076 | 2.53 |
ENSMUST00000034478.3
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr1_-_38875757 | 2.53 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr17_+_70276382 | 2.49 |
ENSMUST00000146730.9
|
Dlgap1
|
DLG associated protein 1 |
| chr17_-_16050913 | 2.49 |
ENSMUST00000231281.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr11_-_86964881 | 2.47 |
ENSMUST00000020804.8
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr9_-_66951114 | 2.45 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr3_-_88280047 | 2.45 |
ENSMUST00000107543.8
ENSMUST00000107542.2 |
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chrX_+_72108393 | 2.41 |
ENSMUST00000060418.8
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr11_-_70924288 | 2.39 |
ENSMUST00000238695.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr2_+_154032731 | 2.35 |
ENSMUST00000081816.11
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chrX_+_135723420 | 2.34 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr7_+_24335969 | 2.34 |
ENSMUST00000080718.6
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr11_-_100026754 | 2.33 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr4_-_155103837 | 2.30 |
ENSMUST00000126098.2
ENSMUST00000176194.8 |
Plch2
|
phospholipase C, eta 2 |
| chr9_+_26645024 | 2.29 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr3_-_94693740 | 2.23 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr17_-_37178079 | 2.22 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr10_+_75009797 | 2.22 |
ENSMUST00000218465.2
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chrX_-_149440388 | 2.22 |
ENSMUST00000151403.9
ENSMUST00000087253.11 ENSMUST00000112709.8 ENSMUST00000163969.8 ENSMUST00000087258.10 |
Tro
|
trophinin |
| chr19_-_10217968 | 2.20 |
ENSMUST00000189897.2
ENSMUST00000186056.7 ENSMUST00000088013.12 |
Myrf
|
myelin regulatory factor |
| chr7_-_121785385 | 2.19 |
ENSMUST00000033153.9
|
Ern2
|
endoplasmic reticulum (ER) to nucleus signalling 2 |
| chrX_-_97934387 | 2.18 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr9_-_66951025 | 2.18 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr11_+_100900278 | 2.17 |
ENSMUST00000103110.10
ENSMUST00000044721.13 ENSMUST00000168757.9 |
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr1_+_132808011 | 2.17 |
ENSMUST00000027706.4
|
Lrrn2
|
leucine rich repeat protein 2, neuronal |
| chrX_-_74621828 | 2.14 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr2_+_103255165 | 2.13 |
ENSMUST00000126290.2
|
Elf5
|
E74-like factor 5 |
| chr17_+_35295909 | 2.13 |
ENSMUST00000013910.5
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr9_-_66951151 | 2.09 |
ENSMUST00000113696.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr4_-_129015027 | 2.09 |
ENSMUST00000030572.10
|
Hpca
|
hippocalcin |
| chr17_-_57289121 | 2.09 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr4_-_148372384 | 2.08 |
ENSMUST00000047720.9
|
Disp3
|
dispatched RND transporter family member 3 |
| chr4_-_57143437 | 2.07 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr14_-_55163452 | 2.06 |
ENSMUST00000227037.2
|
Efs
|
embryonal Fyn-associated substrate |
| chr19_-_5507532 | 2.05 |
ENSMUST00000236881.2
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr14_-_61275340 | 2.03 |
ENSMUST00000225730.2
ENSMUST00000111236.4 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_43139390 | 2.03 |
ENSMUST00000107974.3
|
Iglon5
|
IgLON family member 5 |
| chr12_-_45120895 | 2.03 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr5_-_123270449 | 2.02 |
ENSMUST00000186469.7
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr1_+_34840785 | 2.00 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr15_+_74435217 | 2.00 |
ENSMUST00000190524.7
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr7_+_113365235 | 1.99 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr7_+_49624978 | 1.98 |
ENSMUST00000107603.2
|
Nell1
|
NEL-like 1 |
| chr7_-_97827461 | 1.98 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr16_-_22475915 | 1.97 |
ENSMUST00000089925.10
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr2_+_119067929 | 1.96 |
ENSMUST00000110816.8
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr2_+_119067832 | 1.96 |
ENSMUST00000028783.14
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr15_+_80862074 | 1.95 |
ENSMUST00000229727.2
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr11_+_69920849 | 1.94 |
ENSMUST00000143920.4
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chrX_+_21581135 | 1.94 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr13_-_99653045 | 1.92 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr11_+_69920956 | 1.91 |
ENSMUST00000232115.2
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr4_+_133788065 | 1.88 |
ENSMUST00000227683.2
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr18_-_38342815 | 1.88 |
ENSMUST00000057185.13
|
Pcdh1
|
protocadherin 1 |
| chr4_-_11966367 | 1.87 |
ENSMUST00000056050.5
ENSMUST00000108299.2 ENSMUST00000108297.3 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_+_35295894 | 1.87 |
ENSMUST00000172678.8
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr5_+_57875309 | 1.86 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr18_+_61058716 | 1.86 |
ENSMUST00000115297.8
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_+_76070483 | 1.85 |
ENSMUST00000129853.8
|
Tlcd3a
|
TLC domain containing 3A |
| chr4_-_127247864 | 1.83 |
ENSMUST00000106090.8
ENSMUST00000060419.2 |
Gjb4
|
gap junction protein, beta 4 |
| chr8_-_47866869 | 1.83 |
ENSMUST00000211882.2
|
Stox2
|
storkhead box 2 |
| chr6_-_113696390 | 1.81 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr4_+_138181616 | 1.80 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr18_+_77273510 | 1.80 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr7_+_127347339 | 1.77 |
ENSMUST00000206893.2
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr2_+_28423367 | 1.77 |
ENSMUST00000113893.8
ENSMUST00000100241.10 |
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr9_-_66950991 | 1.76 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr4_+_43381979 | 1.74 |
ENSMUST00000035645.12
ENSMUST00000144911.8 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr10_+_79552421 | 1.74 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr14_-_24054927 | 1.70 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_+_117979899 | 1.69 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr2_+_165345707 | 1.69 |
ENSMUST00000029196.5
|
Slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr5_-_146521629 | 1.68 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr13_-_95661726 | 1.66 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr11_-_101877832 | 1.63 |
ENSMUST00000107173.9
ENSMUST00000107172.8 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr13_+_58956495 | 1.62 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr2_-_173117936 | 1.62 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_182591425 | 1.62 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chrX_-_135116192 | 1.60 |
ENSMUST00000113120.2
ENSMUST00000113118.2 ENSMUST00000058125.9 |
Bex1
|
brain expressed X-linked 1 |
| chr14_-_55163311 | 1.59 |
ENSMUST00000022813.8
|
Efs
|
embryonal Fyn-associated substrate |
| chr4_+_43383449 | 1.59 |
ENSMUST00000135216.2
ENSMUST00000152322.8 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr7_-_103832599 | 1.58 |
ENSMUST00000216612.3
|
Olfr648
|
olfactory receptor 648 |
| chr14_+_70791127 | 1.56 |
ENSMUST00000161069.8
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr4_+_11758147 | 1.56 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr7_+_4928784 | 1.55 |
ENSMUST00000057612.9
|
Ssc5d
|
scavenger receptor cysteine rich family, 5 domains |
| chr9_-_49710190 | 1.54 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_+_21095399 | 1.53 |
ENSMUST00000115458.9
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr12_-_87037204 | 1.53 |
ENSMUST00000222543.2
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr5_-_93192881 | 1.52 |
ENSMUST00000061328.6
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr9_-_44646487 | 1.52 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr1_-_64995891 | 1.49 |
ENSMUST00000123225.2
|
Plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr6_-_137626207 | 1.49 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr10_-_83484467 | 1.46 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr1_-_132294807 | 1.46 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr2_+_102488985 | 1.46 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chrX_+_92718695 | 1.45 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr12_+_84161095 | 1.45 |
ENSMUST00000123491.8
ENSMUST00000046340.9 ENSMUST00000136159.2 |
Dnal1
|
dynein, axonemal, light chain 1 |
| chr2_+_70393782 | 1.45 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr10_+_24099415 | 1.44 |
ENSMUST00000095784.3
|
Moxd1
|
monooxygenase, DBH-like 1 |
| chr3_-_141875070 | 1.42 |
ENSMUST00000106230.2
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr3_-_8732316 | 1.42 |
ENSMUST00000042412.5
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1 |
| chr4_-_46602192 | 1.41 |
ENSMUST00000107756.4
|
Coro2a
|
coronin, actin binding protein 2A |
| chr2_-_150746574 | 1.41 |
ENSMUST00000056149.15
|
Abhd12
|
abhydrolase domain containing 12 |
| chr11_-_97464866 | 1.40 |
ENSMUST00000207653.2
ENSMUST00000107593.8 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr2_-_173118315 | 1.40 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chrX_-_52645649 | 1.40 |
ENSMUST00000088779.5
|
Rtl8a
|
retrotransposon Gag like 8A |
| chr1_+_75377616 | 1.40 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr7_+_127347308 | 1.39 |
ENSMUST00000188580.3
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr16_+_17093941 | 1.39 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr18_+_62681982 | 1.39 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr17_-_16051295 | 1.39 |
ENSMUST00000231985.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr9_-_62444318 | 1.38 |
ENSMUST00000048043.12
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr7_+_141342696 | 1.38 |
ENSMUST00000155534.9
ENSMUST00000041924.14 ENSMUST00000163321.3 |
Muc5ac
|
mucin 5, subtypes A and C, tracheobronchial/gastric |
| chrX_-_149440362 | 1.38 |
ENSMUST00000148604.2
|
Tro
|
trophinin |
| chr18_+_37818263 | 1.37 |
ENSMUST00000194418.2
|
Pcdhga4
|
protocadherin gamma subfamily A, 4 |
| chr2_-_171885386 | 1.33 |
ENSMUST00000087950.4
|
Cbln4
|
cerebellin 4 precursor protein |
| chr11_+_69920542 | 1.32 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr7_-_109322993 | 1.32 |
ENSMUST00000106735.9
ENSMUST00000033334.5 |
BC051019
|
cDNA sequence BC051019 |
| chr8_+_75720286 | 1.32 |
ENSMUST00000211863.2
|
Hmgxb4
|
HMG box domain containing 4 |
| chr9_-_75466865 | 1.32 |
ENSMUST00000215821.2
|
Tmod3
|
tropomodulin 3 |
| chr8_-_59154041 | 1.31 |
ENSMUST00000188531.7
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr4_+_128777339 | 1.31 |
ENSMUST00000035667.9
|
Trim62
|
tripartite motif-containing 62 |
| chr9_-_44632680 | 1.29 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr15_+_80861966 | 1.28 |
ENSMUST00000139517.9
ENSMUST00000137255.3 ENSMUST00000137004.2 |
Sgsm3
|
small G protein signaling modulator 3 |
| chr9_-_58109564 | 1.28 |
ENSMUST00000163897.8
ENSMUST00000215950.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr3_-_132655804 | 1.28 |
ENSMUST00000117164.8
ENSMUST00000093971.5 ENSMUST00000042729.16 |
Npnt
|
nephronectin |
| chr5_+_130200639 | 1.27 |
ENSMUST00000119797.8
ENSMUST00000148264.8 |
Rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chrX_+_73372664 | 1.27 |
ENSMUST00000004326.4
|
Plxna3
|
plexin A3 |
| chr11_-_61470462 | 1.27 |
ENSMUST00000147501.8
ENSMUST00000146455.8 ENSMUST00000108711.8 ENSMUST00000108712.8 ENSMUST00000001063.15 ENSMUST00000108713.8 ENSMUST00000179936.8 ENSMUST00000178202.8 |
Epn2
|
epsin 2 |
| chr3_-_132655954 | 1.26 |
ENSMUST00000042744.16
ENSMUST00000117811.8 |
Npnt
|
nephronectin |
| chr11_+_59197746 | 1.26 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr9_+_26645141 | 1.26 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr13_+_3887757 | 1.25 |
ENSMUST00000042219.6
|
Calm4
|
calmodulin 4 |
| chr2_+_69652714 | 1.25 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr3_+_131270529 | 1.25 |
ENSMUST00000029666.14
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr17_+_28491085 | 1.25 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr6_-_120799500 | 1.24 |
ENSMUST00000204699.2
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr8_-_100143029 | 1.24 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr13_-_34529157 | 1.23 |
ENSMUST00000040336.12
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr8_+_40876827 | 1.23 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr6_+_145879839 | 1.23 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr3_-_94693780 | 1.23 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr6_+_17306334 | 1.21 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr2_-_168123015 | 1.21 |
ENSMUST00000109191.2
|
Kcng1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr9_-_112016966 | 1.21 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr7_+_126396779 | 1.20 |
ENSMUST00000205324.2
|
Tlcd3b
|
TLC domain containing 3B |
| chr3_+_121517158 | 1.20 |
ENSMUST00000029771.13
|
F3
|
coagulation factor III |
| chr8_-_46452896 | 1.19 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 3.1 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 2.0 | 11.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.7 | 11.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 1.3 | 10.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.9 | 2.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.9 | 8.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.8 | 3.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.8 | 2.5 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.8 | 5.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.7 | 3.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 2.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.7 | 9.3 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.7 | 3.3 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.6 | 8.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 3.6 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.6 | 4.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.6 | 1.7 | GO:0070949 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 2.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 1.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.5 | 2.5 | GO:0097195 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.5 | 3.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.5 | 3.0 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.5 | 2.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 1.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.5 | 1.4 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.5 | 1.8 | GO:2000506 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.4 | 1.7 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.4 | 2.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 2.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.4 | 3.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.4 | 1.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.4 | 1.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.4 | 2.7 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.4 | 1.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.4 | 1.1 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.4 | 1.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 | 0.7 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.4 | 4.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.4 | 1.1 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.4 | 2.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.3 | 2.4 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.3 | 2.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 2.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 1.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.3 | 1.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 0.6 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.3 | 1.5 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.3 | 1.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.3 | 7.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 1.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 0.9 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.3 | 1.7 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.3 | 1.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 2.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 5.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 2.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 | 1.6 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.3 | 1.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 2.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 1.0 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 1.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.2 | 1.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 2.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 0.7 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.2 | 0.7 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 | 1.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 0.7 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 2.7 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 4.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 4.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 | 1.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.8 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 0.4 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.2 | 1.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 2.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 3.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 1.1 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 2.8 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 0.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 0.9 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.2 | 2.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 1.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 2.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 8.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 0.5 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.2 | 0.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.8 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 0.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.2 | 2.6 | GO:0061339 | establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.2 | 0.8 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 0.6 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.1 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.9 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 5.8 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 1.4 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.6 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 1.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 4.0 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 1.1 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.1 | 1.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 1.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.3 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.2 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.3 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 1.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.3 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.1 | 1.1 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 2.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 2.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.5 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 2.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.6 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 1.4 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.8 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 2.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 2.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 | 0.3 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.2 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.9 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.7 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.5 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 8.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 2.3 | GO:0061099 | positive regulation of protein tyrosine kinase activity(GO:0061098) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 2.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 2.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 3.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 1.5 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 1.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 4.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 5.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 3.1 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.8 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.6 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 4.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 1.0 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.2 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.8 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 2.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 2.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.5 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 4.7 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.3 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 3.2 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 3.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.3 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.8 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.7 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 1.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 2.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 1.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.5 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 1.6 | GO:0001890 | placenta development(GO:0001890) |
| 0.0 | 1.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.6 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.5 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.4 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.1 | 4.5 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.8 | 11.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.7 | 5.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.7 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 2.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 1.7 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.5 | 3.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 2.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 1.3 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.3 | 0.9 | GO:0097144 | BAX complex(GO:0097144) |
| 0.3 | 2.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 2.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 1.3 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 5.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 7.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 2.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 3.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.6 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 2.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.7 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 3.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 3.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 2.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 16.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 3.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 2.9 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 2.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 4.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 1.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 9.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 3.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 0.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 6.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 5.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 6.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 16.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 3.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 4.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 5.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 7.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 11.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.0 | 3.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.7 | 5.2 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.7 | 2.7 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.7 | 3.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 1.8 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.6 | 3.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.6 | 1.7 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.5 | 1.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.5 | 3.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.5 | 2.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 4.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 1.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.5 | 1.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.4 | 4.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.4 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 1.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 2.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.0 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.3 | 1.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 1.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.3 | 1.3 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.3 | 5.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 1.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 4.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 3.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 6.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 1.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 4.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 2.8 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 0.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 1.6 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 2.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 6.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 1.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 2.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 9.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.9 | GO:0051381 | histamine binding(GO:0051381) |
| 0.2 | 1.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 6.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 11.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 1.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 5.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 1.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 6.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 5.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.5 | GO:0035276 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.1 | 0.6 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 13.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 2.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 9.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.6 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 6.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.4 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.5 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 3.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.2 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 1.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.3 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 2.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 2.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 2.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 2.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 6.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 2.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 3.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 3.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 4.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 7.0 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 2.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) choline binding(GO:0033265) |
| 0.0 | 0.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 3.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 6.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 9.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 5.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 2.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 10.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 5.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 8.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 6.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 8.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 11.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 4.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 4.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 8.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 3.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 4.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 3.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 8.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |