Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Tfap2c
Z-value: 1.07
Transcription factors associated with Tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2c
|
ENSMUSG00000028640.12 | Tfap2c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2c | mm39_v1_chr2_+_172392678_172392702 | 0.09 | 4.4e-01 | Click! |
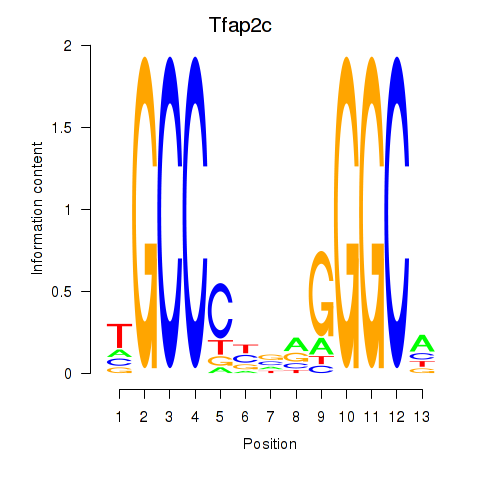
Activity profile of Tfap2c motif
Sorted Z-values of Tfap2c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_10366753 | 9.83 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr5_+_24679154 | 6.05 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr9_+_89791943 | 5.84 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr6_-_42670021 | 5.78 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr6_-_42669963 | 5.67 |
ENSMUST00000045140.5
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr7_-_45019984 | 5.63 |
ENSMUST00000003971.10
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr9_-_121323906 | 5.61 |
ENSMUST00000215228.2
ENSMUST00000213106.2 |
Cck
|
cholecystokinin |
| chr1_-_75240551 | 5.46 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr2_+_156263002 | 5.34 |
ENSMUST00000125153.10
ENSMUST00000103136.8 ENSMUST00000109577.9 |
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr9_+_58395850 | 5.23 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr11_-_119907884 | 5.22 |
ENSMUST00000132575.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr6_+_118043307 | 5.14 |
ENSMUST00000203804.3
ENSMUST00000203482.2 |
Rasgef1a
|
RasGEF domain family, member 1A |
| chr7_-_100543891 | 5.07 |
ENSMUST00000209041.2
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr11_-_65053710 | 5.03 |
ENSMUST00000093002.12
ENSMUST00000047463.15 |
Arhgap44
|
Rho GTPase activating protein 44 |
| chr11_-_94364914 | 4.99 |
ENSMUST00000107786.8
ENSMUST00000107791.8 ENSMUST00000103166.9 ENSMUST00000107792.8 ENSMUST00000100561.10 ENSMUST00000107793.8 ENSMUST00000107788.8 ENSMUST00000107790.8 ENSMUST00000107789.8 ENSMUST00000107785.2 ENSMUST00000021234.15 |
Cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr14_+_7030760 | 4.82 |
ENSMUST00000055211.6
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chrX_-_132589727 | 4.78 |
ENSMUST00000149154.8
|
Pcdh19
|
protocadherin 19 |
| chr11_+_67345895 | 4.77 |
ENSMUST00000108681.9
|
Gas7
|
growth arrest specific 7 |
| chr6_+_38639945 | 4.74 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chr9_-_121825028 | 4.73 |
ENSMUST00000216669.2
ENSMUST00000215084.2 ENSMUST00000214533.2 ENSMUST00000217610.2 ENSMUST00000084743.7 |
Pomgnt2
|
protein O-linked mannose beta 1,4-N-acetylglucosaminyltransferase 2 |
| chr5_-_24806960 | 4.64 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr6_+_88175312 | 4.59 |
ENSMUST00000203480.2
ENSMUST00000015197.9 |
Gata2
|
GATA binding protein 2 |
| chr9_-_20657643 | 4.52 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chrX_+_52609954 | 4.51 |
ENSMUST00000063384.4
|
Rtl8c
|
retrotransposon Gag like 8C |
| chr2_-_32243295 | 4.50 |
ENSMUST00000091089.12
ENSMUST00000078352.12 ENSMUST00000113350.8 ENSMUST00000202578.4 ENSMUST00000113365.8 |
Dnm1
|
dynamin 1 |
| chr1_+_75456173 | 4.48 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr5_-_71253107 | 4.48 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr6_-_28831746 | 4.37 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr4_-_56990306 | 4.31 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr9_-_29323032 | 4.25 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr2_+_35512172 | 4.17 |
ENSMUST00000112992.9
|
Dab2ip
|
disabled 2 interacting protein |
| chr2_+_156262957 | 4.10 |
ENSMUST00000109574.8
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr9_+_121589044 | 4.10 |
ENSMUST00000093772.4
|
Zfp651
|
zinc finger protein 651 |
| chr2_-_170269748 | 4.03 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr18_+_86413077 | 4.02 |
ENSMUST00000058829.4
|
Neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr11_-_59855762 | 3.93 |
ENSMUST00000062405.8
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chr9_+_109760856 | 3.91 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr14_-_65499835 | 3.86 |
ENSMUST00000131309.3
|
Fzd3
|
frizzled class receptor 3 |
| chr2_+_32498997 | 3.84 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr9_-_29323500 | 3.77 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr12_-_81379903 | 3.75 |
ENSMUST00000085238.13
ENSMUST00000182208.8 |
Slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr1_-_84912810 | 3.74 |
ENSMUST00000027422.7
|
Slc16a14
|
solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| chr13_+_54519161 | 3.72 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr14_+_121272606 | 3.67 |
ENSMUST00000135010.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr19_+_10366450 | 3.67 |
ENSMUST00000073899.6
|
Syt7
|
synaptotagmin VII |
| chr7_-_138768374 | 3.66 |
ENSMUST00000016125.12
|
Stk32c
|
serine/threonine kinase 32C |
| chr2_+_156262756 | 3.65 |
ENSMUST00000103137.10
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_-_101875575 | 3.49 |
ENSMUST00000176261.2
ENSMUST00000143177.2 ENSMUST00000003612.13 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr2_-_32243246 | 3.40 |
ENSMUST00000201433.4
ENSMUST00000113352.9 ENSMUST00000201494.4 |
Dnm1
|
dynamin 1 |
| chr2_+_35512023 | 3.38 |
ENSMUST00000091010.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr2_-_32271921 | 3.34 |
ENSMUST00000048792.5
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr15_+_87509413 | 3.29 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr2_+_138098454 | 3.27 |
ENSMUST00000091556.12
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr4_+_124779592 | 3.19 |
ENSMUST00000149146.2
|
Epha10
|
Eph receptor A10 |
| chr7_+_131568167 | 3.16 |
ENSMUST00000045840.5
|
Gpr26
|
G protein-coupled receptor 26 |
| chrX_+_162691978 | 3.09 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_-_75455915 | 3.05 |
ENSMUST00000079205.14
ENSMUST00000094818.4 |
Chpf
|
chondroitin polymerizing factor |
| chr4_-_148244299 | 3.01 |
ENSMUST00000151127.8
ENSMUST00000105705.9 |
Fbxo44
|
F-box protein 44 |
| chrX_-_141089165 | 3.00 |
ENSMUST00000134825.3
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr4_+_138181616 | 2.88 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr12_+_111725357 | 2.88 |
ENSMUST00000118471.8
ENSMUST00000122300.8 |
Klc1
|
kinesin light chain 1 |
| chr9_-_21963306 | 2.85 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr10_-_29411857 | 2.78 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chr15_+_54434576 | 2.76 |
ENSMUST00000025356.4
|
Mal2
|
mal, T cell differentiation protein 2 |
| chr7_-_109559671 | 2.74 |
ENSMUST00000080437.13
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chrX_-_72868544 | 2.70 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr15_-_89033761 | 2.69 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr8_+_26008799 | 2.62 |
ENSMUST00000119398.10
ENSMUST00000117179.9 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr12_+_111725282 | 2.61 |
ENSMUST00000239017.2
ENSMUST00000084941.12 |
Klc1
|
kinesin light chain 1 |
| chr4_+_104224774 | 2.60 |
ENSMUST00000106830.9
|
Dab1
|
disabled 1 |
| chr11_-_28534260 | 2.57 |
ENSMUST00000093253.10
ENSMUST00000109502.9 ENSMUST00000042534.15 |
Ccdc85a
|
coiled-coil domain containing 85A |
| chr11_-_119937896 | 2.56 |
ENSMUST00000064307.10
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr5_+_144482693 | 2.56 |
ENSMUST00000071782.8
|
Nptx2
|
neuronal pentraxin 2 |
| chr3_-_88997261 | 2.54 |
ENSMUST00000196223.5
ENSMUST00000196043.2 ENSMUST00000166687.6 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr14_-_20844034 | 2.53 |
ENSMUST00000226630.2
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chrX_-_20787150 | 2.50 |
ENSMUST00000081893.7
ENSMUST00000115345.8 |
Syn1
|
synapsin I |
| chr11_-_33942981 | 2.49 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr17_-_56783376 | 2.45 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr7_-_109559593 | 2.44 |
ENSMUST00000106722.2
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr8_+_26008773 | 2.41 |
ENSMUST00000084027.13
ENSMUST00000178276.8 ENSMUST00000179592.8 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr10_+_24099415 | 2.40 |
ENSMUST00000095784.3
|
Moxd1
|
monooxygenase, DBH-like 1 |
| chr18_-_33596468 | 2.40 |
ENSMUST00000171533.9
|
Nrep
|
neuronal regeneration related protein |
| chr1_+_181180183 | 2.40 |
ENSMUST00000161880.8
ENSMUST00000027795.14 |
Cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr7_-_140773859 | 2.38 |
ENSMUST00000026572.11
ENSMUST00000168550.8 ENSMUST00000097957.11 |
Hras
|
Harvey rat sarcoma virus oncogene |
| chr19_-_6964988 | 2.37 |
ENSMUST00000130048.8
ENSMUST00000025914.7 |
Vegfb
|
vascular endothelial growth factor B |
| chr9_-_107109108 | 2.36 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr4_+_48585135 | 2.36 |
ENSMUST00000030032.13
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr2_+_174169539 | 2.33 |
ENSMUST00000133356.8
ENSMUST00000087871.11 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr17_-_56783462 | 2.32 |
ENSMUST00000067538.6
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr2_+_35146390 | 2.31 |
ENSMUST00000201185.4
ENSMUST00000202990.4 ENSMUST00000202899.4 ENSMUST00000142324.8 ENSMUST00000139867.5 |
Gsn
|
gelsolin |
| chr4_+_125384481 | 2.30 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr14_+_121272950 | 2.30 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr11_+_98274637 | 2.29 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr17_+_37357451 | 2.28 |
ENSMUST00000172789.2
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr4_+_152423075 | 2.27 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr2_+_174169351 | 2.26 |
ENSMUST00000124935.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr4_+_48585275 | 2.23 |
ENSMUST00000123476.8
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr2_-_165076609 | 2.22 |
ENSMUST00000065438.13
|
Cdh22
|
cadherin 22 |
| chr2_-_32271833 | 2.22 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr7_-_19338349 | 2.21 |
ENSMUST00000086041.7
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chrX_-_100128958 | 2.20 |
ENSMUST00000101362.8
ENSMUST00000073927.5 |
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr6_+_4504814 | 2.20 |
ENSMUST00000141483.8
|
Col1a2
|
collagen, type I, alpha 2 |
| chr14_+_119375753 | 2.19 |
ENSMUST00000065904.5
|
Hs6st3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chrX_-_47123719 | 2.19 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr16_+_91022300 | 2.16 |
ENSMUST00000035608.10
|
Olig2
|
oligodendrocyte transcription factor 2 |
| chrX_+_162692126 | 2.12 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_+_102080489 | 2.11 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr2_+_174169492 | 2.09 |
ENSMUST00000156623.8
ENSMUST00000149016.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr18_-_61147272 | 2.02 |
ENSMUST00000025520.10
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chr11_-_80268800 | 2.01 |
ENSMUST00000179332.8
ENSMUST00000103225.11 ENSMUST00000134274.2 |
5730455P16Rik
|
RIKEN cDNA 5730455P16 gene |
| chr19_+_5118103 | 2.01 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr19_+_10019023 | 2.00 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr4_+_152423344 | 1.98 |
ENSMUST00000005175.5
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr3_-_154302679 | 1.97 |
ENSMUST00000052774.8
ENSMUST00000170461.8 ENSMUST00000122976.2 |
Tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr10_-_75396164 | 1.97 |
ENSMUST00000051129.10
|
Lrrc75b
|
leucine rich repeat containing 75B |
| chr12_-_65120674 | 1.95 |
ENSMUST00000220983.2
ENSMUST00000220730.2 ENSMUST00000021332.10 |
Fkbp3
|
FK506 binding protein 3 |
| chr2_+_139520098 | 1.93 |
ENSMUST00000184404.8
ENSMUST00000099307.4 |
Ism1
|
isthmin 1, angiogenesis inhibitor |
| chr4_-_96029375 | 1.92 |
ENSMUST00000097972.5
|
Cyp2j12
|
cytochrome P450, family 2, subfamily j, polypeptide 12 |
| chr7_-_43139390 | 1.88 |
ENSMUST00000107974.3
|
Iglon5
|
IgLON family member 5 |
| chrX_+_71006170 | 1.87 |
ENSMUST00000126362.8
ENSMUST00000170096.8 |
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr17_-_16050913 | 1.86 |
ENSMUST00000231281.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr12_-_76756772 | 1.86 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr18_-_62044871 | 1.82 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr14_+_120715855 | 1.82 |
ENSMUST00000062117.14
|
Rap2a
|
RAS related protein 2a |
| chr5_-_35836761 | 1.81 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr17_+_86475205 | 1.80 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr11_+_102080446 | 1.79 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr4_+_45203914 | 1.79 |
ENSMUST00000107804.2
|
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr11_+_70431063 | 1.79 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr2_+_135501605 | 1.75 |
ENSMUST00000134310.8
|
Plcb4
|
phospholipase C, beta 4 |
| chr17_-_30107544 | 1.72 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr6_-_32565127 | 1.68 |
ENSMUST00000115096.4
|
Plxna4
|
plexin A4 |
| chrX_+_133657312 | 1.67 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr18_-_33596890 | 1.64 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr4_+_40473130 | 1.63 |
ENSMUST00000179526.2
|
Tmem215
|
transmembrane protein 215 |
| chr17_-_66756710 | 1.62 |
ENSMUST00000086693.12
ENSMUST00000097291.10 |
Mtcl1
|
microtubule crosslinking factor 1 |
| chr10_-_13744523 | 1.62 |
ENSMUST00000105534.10
|
Aig1
|
androgen-induced 1 |
| chr3_+_154302989 | 1.60 |
ENSMUST00000140644.8
ENSMUST00000144764.8 ENSMUST00000155232.2 |
Cryz
|
crystallin, zeta |
| chr7_-_16550656 | 1.60 |
ENSMUST00000061390.9
|
Fkrp
|
fukutin related protein |
| chr19_+_59446804 | 1.58 |
ENSMUST00000062216.4
|
Emx2
|
empty spiracles homeobox 2 |
| chr4_+_131649001 | 1.57 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr9_-_42035560 | 1.56 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr9_+_108174052 | 1.54 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr8_-_37081091 | 1.53 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr6_+_40302106 | 1.53 |
ENSMUST00000031977.12
|
Agk
|
acylglycerol kinase |
| chr10_-_121146940 | 1.52 |
ENSMUST00000064107.7
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr9_+_74944994 | 1.51 |
ENSMUST00000007800.8
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_-_58346806 | 1.50 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr10_-_13744676 | 1.50 |
ENSMUST00000019942.6
ENSMUST00000162610.8 |
Aig1
|
androgen-induced 1 |
| chr7_-_138447933 | 1.50 |
ENSMUST00000118810.2
ENSMUST00000075667.5 ENSMUST00000119664.2 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr11_-_97590460 | 1.49 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr4_-_148021159 | 1.48 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr11_+_77384234 | 1.46 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr5_-_114582053 | 1.45 |
ENSMUST00000123256.3
ENSMUST00000112245.6 |
Mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human) |
| chr4_-_148021217 | 1.44 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr2_+_103788321 | 1.44 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr7_+_6289572 | 1.44 |
ENSMUST00000086327.12
ENSMUST00000153840.2 ENSMUST00000170776.8 |
Zfp667
|
zinc finger protein 667 |
| chr6_-_12749192 | 1.42 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr6_-_97037366 | 1.41 |
ENSMUST00000089295.6
|
Tafa4
|
TAFA chemokine like family member 4 |
| chr11_-_76737794 | 1.41 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr11_-_66416621 | 1.38 |
ENSMUST00000123454.8
|
Shisa6
|
shisa family member 6 |
| chr8_+_84699580 | 1.36 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr9_+_45341589 | 1.34 |
ENSMUST00000239471.2
ENSMUST00000034592.11 ENSMUST00000239429.2 |
Dscaml1
|
DS cell adhesion molecule like 1 |
| chrX_+_128650486 | 1.33 |
ENSMUST00000167619.9
ENSMUST00000037854.15 |
Diaph2
|
diaphanous related formin 2 |
| chrX_+_71006577 | 1.31 |
ENSMUST00000048790.7
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr11_+_78389913 | 1.30 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr9_-_105705321 | 1.29 |
ENSMUST00000098441.10
|
Col6a6
|
collagen, type VI, alpha 6 |
| chr3_+_154302777 | 1.27 |
ENSMUST00000194876.6
|
Cryz
|
crystallin, zeta |
| chr4_-_116321348 | 1.25 |
ENSMUST00000106486.8
ENSMUST00000106485.8 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr17_-_46867083 | 1.22 |
ENSMUST00000015749.7
|
Srf
|
serum response factor |
| chr3_+_154302311 | 1.22 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr2_+_81883566 | 1.21 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr6_+_22288220 | 1.21 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr15_+_80171435 | 1.21 |
ENSMUST00000160424.8
|
Cacna1i
|
calcium channel, voltage-dependent, alpha 1I subunit |
| chr18_+_65020910 | 1.18 |
ENSMUST00000236736.2
ENSMUST00000235743.2 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr18_-_33597060 | 1.17 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr2_+_29759495 | 1.16 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr3_-_107993906 | 1.16 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr9_+_109760931 | 1.15 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr7_+_16186704 | 1.14 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr18_-_33596792 | 1.13 |
ENSMUST00000051087.16
|
Nrep
|
neuronal regeneration related protein |
| chr3_-_148696155 | 1.12 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr19_+_10018914 | 1.11 |
ENSMUST00000115995.4
|
Fads3
|
fatty acid desaturase 3 |
| chr9_+_109760528 | 1.10 |
ENSMUST00000035055.15
|
Map4
|
microtubule-associated protein 4 |
| chr5_-_114582097 | 1.10 |
ENSMUST00000031560.14
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human) |
| chr17_-_16051295 | 1.08 |
ENSMUST00000231985.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr4_+_105014536 | 1.07 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr7_+_65511482 | 1.05 |
ENSMUST00000176199.8
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr7_+_3697511 | 1.05 |
ENSMUST00000108627.4
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr4_-_117146624 | 1.04 |
ENSMUST00000221654.2
|
Rnf220
|
ring finger protein 220 |
| chr5_-_92582969 | 1.04 |
ENSMUST00000135112.8
|
Nup54
|
nucleoporin 54 |
| chr5_+_137756407 | 1.03 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr18_+_65020760 | 1.01 |
ENSMUST00000235343.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr11_+_70430870 | 1.00 |
ENSMUST00000157075.8
|
Pld2
|
phospholipase D2 |
| chr10_-_127177729 | 0.99 |
ENSMUST00000026474.5
ENSMUST00000219671.2 |
Gli1
|
GLI-Kruppel family member GLI1 |
| chr11_+_85243970 | 0.99 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr13_-_72111832 | 0.98 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr9_+_74945021 | 0.96 |
ENSMUST00000168301.2
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr8_-_65302573 | 0.96 |
ENSMUST00000210166.2
|
Klhl2
|
kelch-like 2, Mayven |
| chr9_-_39918243 | 0.96 |
ENSMUST00000073932.4
|
Olfr980
|
olfactory receptor 980 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.7 | 5.0 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 1.5 | 7.5 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 1.4 | 11.5 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 1.3 | 7.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.1 | 4.6 | GO:0035854 | eosinophil fate commitment(GO:0035854) |
| 1.1 | 5.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.1 | 5.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.0 | 5.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.0 | 4.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.0 | 3.9 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 1.0 | 4.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.9 | 5.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.9 | 6.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 2.6 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.8 | 2.4 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.7 | 6.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.7 | 2.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.7 | 2.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.7 | 4.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.6 | 7.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.6 | 2.9 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.6 | 1.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 2.3 | GO:1902174 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.6 | 6.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.5 | 2.7 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.5 | 1.6 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.5 | 3.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 4.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.5 | 6.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 2.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.5 | 2.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.4 | 2.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 1.6 | GO:2001137 | protein retention in Golgi apparatus(GO:0045053) positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 3.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 5.6 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.4 | 2.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.4 | 2.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.8 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 2.8 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 2.7 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 1.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 2.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 2.3 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.3 | 1.9 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.3 | 5.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 0.9 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.3 | 1.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 4.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 2.4 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.3 | 3.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 1.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 5.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 1.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 2.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 1.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 13.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.2 | 3.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 3.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.4 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 2.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 2.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.9 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 4.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 1.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.1 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.2 | 1.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 4.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 0.8 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.2 | 0.8 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 3.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 0.8 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 0.9 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.2 | 2.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 2.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 4.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 1.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 1.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 3.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.4 | GO:1900106 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 3.9 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.6 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 2.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.6 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.1 | 0.5 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 6.7 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 2.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 1.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 4.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.1 | 1.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 2.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 2.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.6 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.1 | 1.5 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 1.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 5.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 2.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 1.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.2 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.4 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 10.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 5.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 4.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 4.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 1.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.9 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 2.9 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.8 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 2.6 | GO:0008306 | associative learning(GO:0008306) |
| 0.0 | 4.8 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 4.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.9 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.8 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 1.4 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 3.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 1.0 | 2.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.9 | 13.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.7 | 3.7 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.7 | 5.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 2.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 9.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 5.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.3 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 7.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 4.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 2.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 5.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 11.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 6.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 5.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 4.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 7.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.8 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 1.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 5.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 7.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 38.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 4.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.8 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 6.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 3.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 3.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 4.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 4.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 5.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 5.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.2 | 3.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 1.1 | 4.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 1.0 | 4.9 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.0 | 3.9 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.9 | 7.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.8 | 5.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.8 | 2.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.8 | 2.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.7 | 2.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 7.9 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.6 | 1.8 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.6 | 2.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.6 | 2.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 2.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.5 | 2.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.5 | 6.7 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 1.5 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.5 | 1.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.5 | 2.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 5.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 4.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 2.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 3.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 4.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 3.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 2.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 2.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 2.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 1.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 1.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 2.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 4.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 2.8 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 2.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 3.5 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 1.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 1.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 2.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 6.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 3.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 5.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 12.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 3.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 1.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.9 | GO:0008823 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 4.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 14.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 6.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 4.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 7.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 6.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 4.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 5.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 4.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 3.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 8.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 5.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 3.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.6 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 7.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 3.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 5.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 2.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 2.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 2.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 12.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 10.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 10.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 5.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 12.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 11.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 5.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 5.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 8.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 5.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 13.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.3 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 5.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 3.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 4.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 2.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 1.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 5.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 3.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 4.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 5.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 2.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 5.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.2 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 4.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 5.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 7.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 4.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |