Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
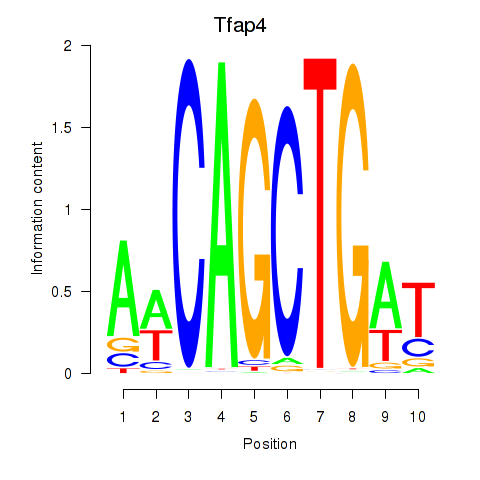
Results for Tfap4
Z-value: 1.65
Transcription factors associated with Tfap4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap4
|
ENSMUSG00000005718.9 | Tfap4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap4 | mm39_v1_chr16_-_4377640_4377718 | -0.22 | 6.1e-02 | Click! |
Activity profile of Tfap4 motif
Sorted Z-values of Tfap4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_58884038 | 14.44 |
ENSMUST00000059539.5
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr7_-_119801327 | 14.15 |
ENSMUST00000033198.6
|
Crym
|
crystallin, mu |
| chr10_+_127337541 | 11.53 |
ENSMUST00000160019.8
ENSMUST00000160610.2 ENSMUST00000035839.3 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr18_+_82572595 | 10.92 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr13_+_46571910 | 9.96 |
ENSMUST00000037923.5
|
Rbm24
|
RNA binding motif protein 24 |
| chr6_+_107506678 | 9.74 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr6_-_36787096 | 9.59 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr15_-_99425555 | 9.54 |
ENSMUST00000231171.2
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr8_+_13034245 | 8.12 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr7_-_126046814 | 7.96 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr2_+_97298002 | 7.56 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr8_-_37200051 | 7.31 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr11_+_102727122 | 7.11 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr9_+_75093177 | 7.06 |
ENSMUST00000129281.8
ENSMUST00000148144.8 ENSMUST00000130384.2 |
Myo5a
|
myosin VA |
| chr5_-_73805063 | 7.02 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr2_-_148574353 | 6.98 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr9_+_121606750 | 6.74 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr6_-_115228800 | 6.73 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr13_+_93444514 | 6.72 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_159351337 | 6.54 |
ENSMUST00000192069.6
|
Tnr
|
tenascin R |
| chr2_-_80277969 | 6.44 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr12_+_88689638 | 6.39 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr6_-_54543446 | 6.39 |
ENSMUST00000019268.11
|
Scrn1
|
secernin 1 |
| chr8_+_55003359 | 6.34 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr2_-_92222979 | 6.29 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chrX_+_100492684 | 6.27 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr13_+_55097200 | 5.98 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr6_-_92458324 | 5.97 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr17_+_27171834 | 5.74 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr1_-_165955445 | 5.72 |
ENSMUST00000085992.4
ENSMUST00000192369.6 |
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr19_-_40502117 | 5.64 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr2_-_52225146 | 5.58 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr9_-_79884920 | 5.55 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr7_+_126811831 | 5.53 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_+_143838149 | 5.51 |
ENSMUST00000146006.3
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr13_-_32967937 | 5.51 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr2_-_52225763 | 5.48 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr19_-_5560473 | 5.48 |
ENSMUST00000237111.2
ENSMUST00000070172.6 |
Snx32
|
sorting nexin 32 |
| chr19_-_8816530 | 5.39 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr5_+_91665474 | 5.29 |
ENSMUST00000040576.10
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr9_-_79885063 | 5.24 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr18_-_60724855 | 5.20 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr13_+_93441447 | 5.16 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr11_-_68762664 | 5.10 |
ENSMUST00000101017.9
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr1_+_171077984 | 5.03 |
ENSMUST00000111315.9
ENSMUST00000219033.2 |
Adamts4
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 4 |
| chr5_-_103174794 | 5.00 |
ENSMUST00000128869.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr3_+_84832783 | 4.97 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr3_-_108062172 | 4.92 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr8_+_55024446 | 4.83 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr18_-_89787603 | 4.82 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr7_+_4925781 | 4.74 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr2_-_74409225 | 4.70 |
ENSMUST00000134168.2
ENSMUST00000111993.9 ENSMUST00000064503.13 |
Lnpk
|
lunapark, ER junction formation factor |
| chr2_+_155224105 | 4.69 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr5_+_14564932 | 4.68 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr11_+_110858842 | 4.67 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_+_112436466 | 4.63 |
ENSMUST00000075477.8
|
Cav3
|
caveolin 3 |
| chr8_-_71257623 | 4.61 |
ENSMUST00000212875.2
ENSMUST00000212001.2 ENSMUST00000212757.2 |
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr14_+_123897383 | 4.60 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chr1_-_97904958 | 4.60 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_-_84970617 | 4.56 |
ENSMUST00000226232.2
ENSMUST00000227021.2 ENSMUST00000005406.12 ENSMUST00000227723.2 |
App
|
amyloid beta (A4) precursor protein |
| chr3_-_84128160 | 4.53 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr16_+_20408886 | 4.53 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr13_-_8921732 | 4.42 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr17_-_43813664 | 4.42 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr2_-_74409723 | 4.29 |
ENSMUST00000130586.8
|
Lnpk
|
lunapark, ER junction formation factor |
| chr14_+_70768257 | 4.26 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr1_-_14380418 | 4.25 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_-_119038162 | 4.20 |
ENSMUST00000084797.6
|
Slc22a13
|
solute carrier family 22 (organic cation transporter), member 13 |
| chr9_+_121539395 | 4.19 |
ENSMUST00000035113.11
ENSMUST00000215966.2 ENSMUST00000215833.2 ENSMUST00000215104.2 |
Ss18l2
|
SS18, nBAF chromatin remodeling complex subunit like 2 |
| chrX_-_47123719 | 4.12 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chrX_+_168468186 | 4.11 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr7_+_107166653 | 4.09 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr5_-_147662798 | 4.09 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr15_-_102630496 | 4.03 |
ENSMUST00000171838.2
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr9_-_121621544 | 4.00 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chrX_+_55825033 | 3.99 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr2_+_121125918 | 3.98 |
ENSMUST00000110639.8
|
Map1a
|
microtubule-associated protein 1 A |
| chr12_-_78908409 | 3.95 |
ENSMUST00000021536.9
|
Atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr18_+_57601541 | 3.92 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr18_+_38429792 | 3.86 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr7_+_143838191 | 3.84 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_+_38430205 | 3.84 |
ENSMUST00000235491.2
|
Rnf14
|
ring finger protein 14 |
| chr2_+_170573727 | 3.82 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
| chr8_-_47742429 | 3.82 |
ENSMUST00000079195.6
|
Stox2
|
storkhead box 2 |
| chr6_+_126916919 | 3.81 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr2_+_155223728 | 3.81 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr6_+_21215472 | 3.76 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr14_+_67470884 | 3.72 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr7_-_141015240 | 3.70 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr10_+_24025188 | 3.67 |
ENSMUST00000020174.7
ENSMUST00000220041.2 |
Stx7
|
syntaxin 7 |
| chr5_-_116221293 | 3.67 |
ENSMUST00000111997.3
ENSMUST00000239438.2 |
Tmem233
|
transmembrane protein 233 |
| chrX_+_55824797 | 3.64 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr2_+_174602412 | 3.63 |
ENSMUST00000029030.9
|
Edn3
|
endothelin 3 |
| chr6_-_53797748 | 3.62 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr14_+_70768289 | 3.61 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr10_-_102326286 | 3.61 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
| chr16_-_84970580 | 3.58 |
ENSMUST00000227737.2
ENSMUST00000226801.2 |
App
|
amyloid beta (A4) precursor protein |
| chr19_+_34585322 | 3.51 |
ENSMUST00000076249.6
|
Ifit3b
|
interferon-induced protein with tetratricopeptide repeats 3B |
| chr1_-_14380327 | 3.51 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_-_107607343 | 3.46 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr8_-_84299540 | 3.46 |
ENSMUST00000212990.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr11_+_109376432 | 3.45 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr13_-_47168286 | 3.45 |
ENSMUST00000052747.4
|
Nhlrc1
|
NHL repeat containing 1 |
| chr5_+_31274064 | 3.44 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr2_-_73410632 | 3.43 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr10_+_69816153 | 3.41 |
ENSMUST00000182207.8
|
Ank3
|
ankyrin 3, epithelial |
| chr8_+_15107646 | 3.37 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr1_+_128069677 | 3.34 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr14_-_45456821 | 3.32 |
ENSMUST00000128484.8
ENSMUST00000147853.8 ENSMUST00000022377.11 ENSMUST00000143609.2 ENSMUST00000153383.8 ENSMUST00000139526.9 |
Txndc16
|
thioredoxin domain containing 16 |
| chr11_+_102159558 | 3.32 |
ENSMUST00000036467.5
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr3_-_116047148 | 3.31 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr5_+_134128543 | 3.31 |
ENSMUST00000016088.9
|
Castor2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr6_+_126916956 | 3.29 |
ENSMUST00000201617.2
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr2_+_106523532 | 3.28 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr16_-_89615225 | 3.27 |
ENSMUST00000164263.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr8_-_84299850 | 3.19 |
ENSMUST00000212703.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr8_+_66838927 | 3.19 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr18_+_38429688 | 3.19 |
ENSMUST00000170811.8
ENSMUST00000072376.13 ENSMUST00000237903.2 ENSMUST00000236116.2 ENSMUST00000237824.2 ENSMUST00000236982.2 ENSMUST00000235549.2 ENSMUST00000237416.2 ENSMUST00000237089.2 |
Rnf14
|
ring finger protein 14 |
| chr18_-_39062201 | 3.13 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr4_-_134431534 | 3.13 |
ENSMUST00000054096.13
ENSMUST00000038628.4 |
Man1c1
|
mannosidase, alpha, class 1C, member 1 |
| chr5_+_102872838 | 3.12 |
ENSMUST00000112853.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr8_-_106198112 | 3.09 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr10_+_80136830 | 3.09 |
ENSMUST00000140828.8
ENSMUST00000138909.8 |
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr5_+_31274046 | 3.06 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr9_-_40257586 | 3.03 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr16_-_88087087 | 3.03 |
ENSMUST00000211444.2
ENSMUST00000023652.16 ENSMUST00000072256.13 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr19_+_34560922 | 3.02 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_-_80642969 | 3.01 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr7_-_97066937 | 3.00 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chrX_-_141089165 | 2.98 |
ENSMUST00000134825.3
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr14_+_3575406 | 2.97 |
ENSMUST00000124353.2
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr18_+_38429858 | 2.94 |
ENSMUST00000171461.3
ENSMUST00000235811.2 |
Rnf14
|
ring finger protein 14 |
| chr9_-_44162762 | 2.91 |
ENSMUST00000034618.6
|
Pdzd3
|
PDZ domain containing 3 |
| chr6_+_47021979 | 2.89 |
ENSMUST00000150737.2
|
Cntnap2
|
contactin associated protein-like 2 |
| chr5_-_146521629 | 2.89 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr10_+_39296005 | 2.87 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr3_+_90444537 | 2.86 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr1_-_25267894 | 2.83 |
ENSMUST00000126626.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr8_+_45960931 | 2.82 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_-_76906832 | 2.81 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr14_+_54496683 | 2.76 |
ENSMUST00000041197.13
ENSMUST00000239403.2 ENSMUST00000197605.3 |
Abhd4
|
abhydrolase domain containing 4 |
| chr17_+_14087827 | 2.75 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr11_-_12362136 | 2.75 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr13_+_64309675 | 2.70 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr19_+_45035942 | 2.67 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr11_+_68582923 | 2.65 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr5_-_137246852 | 2.65 |
ENSMUST00000179412.2
|
Muc3a
|
mucin 3A, cell surface associated |
| chr5_+_28047147 | 2.63 |
ENSMUST00000036227.7
|
Htr5a
|
5-hydroxytryptamine (serotonin) receptor 5A |
| chr11_+_68582897 | 2.62 |
ENSMUST00000108673.8
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr6_+_34840057 | 2.60 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr8_+_123912976 | 2.58 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr6_+_90442269 | 2.58 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr7_-_4517559 | 2.57 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr7_+_105290259 | 2.55 |
ENSMUST00000209445.2
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr16_+_48662894 | 2.55 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr8_+_84699580 | 2.54 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr18_+_38430015 | 2.53 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chr15_-_102630589 | 2.51 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr8_+_83891972 | 2.49 |
ENSMUST00000034145.11
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr16_+_16844217 | 2.45 |
ENSMUST00000232067.2
|
Mapk1
|
mitogen-activated protein kinase 1 |
| chr4_-_43040278 | 2.40 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr6_-_131655849 | 2.40 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr14_+_67470735 | 2.40 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr5_-_131567526 | 2.40 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr3_-_89230190 | 2.38 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr15_+_38662158 | 2.37 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr10_-_7539333 | 2.30 |
ENSMUST00000162606.8
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr5_-_137246611 | 2.27 |
ENSMUST00000196391.5
|
Muc3a
|
mucin 3A, cell surface associated |
| chrX_+_92718695 | 2.25 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_+_110858882 | 2.23 |
ENSMUST00000125692.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_-_50605079 | 2.22 |
ENSMUST00000120622.2
|
Dixdc1
|
DIX domain containing 1 |
| chr14_+_20757615 | 2.19 |
ENSMUST00000022358.9
ENSMUST00000224751.2 |
Zswim8
|
zinc finger SWIM-type containing 8 |
| chrX_+_139565657 | 2.18 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr7_+_15832383 | 2.18 |
ENSMUST00000006181.7
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chr15_-_97665524 | 2.15 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr11_+_97253221 | 2.14 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chr1_-_80643024 | 2.12 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr2_-_104542467 | 2.11 |
ENSMUST00000111118.8
ENSMUST00000028597.4 |
Tcp11l1
|
t-complex 11 like 1 |
| chr3_-_51468236 | 2.10 |
ENSMUST00000037141.9
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr6_+_8259405 | 2.09 |
ENSMUST00000160705.8
ENSMUST00000159433.8 |
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr5_+_117495337 | 2.07 |
ENSMUST00000031309.16
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr6_+_34840151 | 2.07 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr15_+_80171435 | 2.07 |
ENSMUST00000160424.8
|
Cacna1i
|
calcium channel, voltage-dependent, alpha 1I subunit |
| chr8_-_47742389 | 2.06 |
ENSMUST00000211737.2
|
Stox2
|
storkhead box 2 |
| chr4_+_118522716 | 2.05 |
ENSMUST00000102666.5
|
Olfr62
|
olfactory receptor 62 |
| chrX_+_133305529 | 2.04 |
ENSMUST00000113224.9
ENSMUST00000113226.2 |
Drp2
|
dystrophin related protein 2 |
| chr5_-_69499014 | 1.90 |
ENSMUST00000087231.4
ENSMUST00000054095.6 |
Kctd8
|
potassium channel tetramerisation domain containing 8 |
| chr1_+_187730032 | 1.90 |
ENSMUST00000110938.2
|
Esrrg
|
estrogen-related receptor gamma |
| chr15_-_97665679 | 1.90 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chrX_+_9751861 | 1.89 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr1_-_162485675 | 1.89 |
ENSMUST00000193898.2
ENSMUST00000169439.3 |
Myocos
|
myocilin opposite strand |
| chr7_-_4517608 | 1.88 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr18_+_73996743 | 1.88 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr15_-_75620060 | 1.84 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr4_+_152270636 | 1.82 |
ENSMUST00000030779.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr8_+_75760301 | 1.82 |
ENSMUST00000165630.3
ENSMUST00000212651.2 ENSMUST00000212388.2 ENSMUST00000212299.2 ENSMUST00000078847.13 ENSMUST00000211869.2 |
Tom1
|
target of myb1 trafficking protein |
| chr3_+_90444613 | 1.80 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr8_+_3550450 | 1.79 |
ENSMUST00000004683.13
ENSMUST00000160338.2 |
Mcoln1
|
mucolipin 1 |
| chr10_+_60113449 | 1.79 |
ENSMUST00000105465.8
ENSMUST00000179238.8 ENSMUST00000177779.8 ENSMUST00000004316.15 |
Psap
|
prosaposin |
| chr4_+_48045143 | 1.77 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 2.7 | 8.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 2.7 | 8.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 2.4 | 9.6 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.8 | 14.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.6 | 11.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.6 | 4.7 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.4 | 7.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.4 | 4.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.3 | 4.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 1.3 | 5.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.3 | 6.4 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 1.2 | 6.0 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 1.1 | 6.5 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.1 | 3.3 | GO:1904268 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 1.1 | 3.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 1.0 | 4.0 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 1.0 | 9.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.0 | 5.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.0 | 6.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.9 | 3.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.9 | 2.6 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.9 | 6.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.9 | 5.2 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.9 | 2.6 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.8 | 9.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.8 | 4.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 5.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.7 | 4.6 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.7 | 9.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.6 | 2.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.6 | 2.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.6 | 11.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.6 | 4.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.5 | 0.5 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.5 | 6.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 2.7 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.5 | 2.6 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.5 | 4.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.5 | 5.1 | GO:0051081 | membrane disassembly(GO:0030397) positive regulation of axon regeneration(GO:0048680) nuclear envelope disassembly(GO:0051081) |
| 0.5 | 6.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.5 | 7.0 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.5 | 2.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.5 | 9.5 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.5 | 1.4 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.4 | 3.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.8 | GO:1900625 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 2.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 7.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 2.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.4 | 3.0 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.4 | 1.5 | GO:1903923 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.4 | 1.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 16.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.3 | 3.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 2.2 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 | 2.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 3.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 11.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 0.9 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 4.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 2.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 11.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 0.9 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 3.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 4.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 3.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.3 | 1.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 6.7 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.3 | 2.4 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.3 | 2.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.3 | 1.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 6.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 1.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 1.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 6.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 3.8 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.2 | 11.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 2.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 1.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 3.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 3.8 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 | 8.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 2.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 17.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.2 | 2.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.4 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.1 | 7.1 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 5.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 12.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 5.7 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 9.4 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.1 | 0.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 7.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.7 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 2.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 3.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 3.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 1.8 | GO:0060736 | prostate gland growth(GO:0060736) epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 3.4 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 4.0 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 2.6 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 3.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 9.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 3.5 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 | 0.5 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 2.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.5 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 3.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.1 | 1.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 5.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 2.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 1.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.1 | 4.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 9.8 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.4 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 1.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 5.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.4 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 8.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.5 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 1.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 2.3 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 4.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 3.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 5.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 2.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 1.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.5 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.2 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 2.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 3.6 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.8 | 7.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.6 | 6.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.6 | 4.7 | GO:0044317 | rod spherule(GO:0044317) |
| 1.5 | 9.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.4 | 10.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.3 | 5.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.2 | 5.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.0 | 4.0 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 1.0 | 8.0 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 5.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.8 | 7.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 5.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 8.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.7 | 9.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 2.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.5 | 3.7 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 4.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.5 | 3.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 2.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 3.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.4 | 1.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 2.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 4.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 15.0 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 5.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 4.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 3.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 9.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 3.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 12.9 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 1.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 6.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 9.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 0.9 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.2 | 3.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 15.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 3.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 6.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 11.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 15.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 7.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 6.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 9.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 5.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 4.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 13.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 10.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 9.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 1.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 16.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 6.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 9.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 16.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 7.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 9.8 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 4.0 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 5.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.5 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.8 | 9.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.3 | 5.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.1 | 4.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.1 | 14.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.1 | 3.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.0 | 5.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.0 | 5.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 1.0 | 8.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.0 | 14.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.0 | 5.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.9 | 5.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.8 | 5.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.8 | 3.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.8 | 3.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.7 | 6.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.7 | 10.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 5.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 8.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.6 | 4.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 2.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 2.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 2.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 3.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 1.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.5 | 4.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 2.9 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.4 | 7.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 9.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 1.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 6.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.4 | 2.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 4.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 6.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 2.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 4.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 6.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 16.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 1.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 6.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 2.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 3.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 4.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 6.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 3.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.9 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 1.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 3.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 4.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 6.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 3.0 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 5.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 7.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 2.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 3.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 13.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 3.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 1.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 1.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 3.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 1.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 3.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 3.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 4.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 8.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 6.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 3.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 3.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 5.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 5.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 5.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 9.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 6.7 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 10.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 3.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 9.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 9.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 9.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 3.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 10.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 4.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 3.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 4.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 3.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 5.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 2.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 9.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 3.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 4.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 13.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 4.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 3.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 9.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 13.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 8.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 9.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 7.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 11.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 4.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 5.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 18.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 2.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 15.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 7.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 4.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 3.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 6.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 7.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 8.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 7.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 3.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 16.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 13.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 3.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 6.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 6.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 6.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 3.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 5.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 17.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 7.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 4.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 3.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |