Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
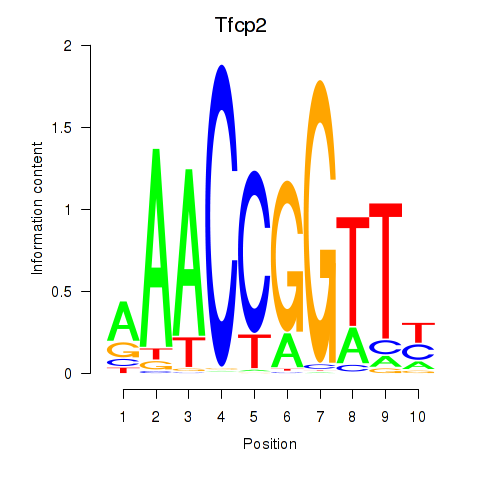
Results for Tfcp2
Z-value: 1.57
Transcription factors associated with Tfcp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2
|
ENSMUSG00000009733.10 | Tfcp2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2 | mm39_v1_chr15_-_100449839_100449892 | 0.05 | 7.0e-01 | Click! |
Activity profile of Tfcp2 motif
Sorted Z-values of Tfcp2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_43455919 | 20.13 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen |
| chr11_+_96820091 | 18.73 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr10_+_43455157 | 14.97 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr4_+_114945905 | 11.58 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr4_-_133329479 | 11.49 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr4_-_116024788 | 10.05 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chrX_+_55824797 | 9.18 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr8_+_57964956 | 8.29 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr8_+_57964921 | 7.97 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr16_-_19079594 | 7.90 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr11_-_95805710 | 7.50 |
ENSMUST00000038343.7
|
B4galnt2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
| chr19_-_34143437 | 7.33 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chrX_-_111608339 | 7.31 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr1_+_44590643 | 6.86 |
ENSMUST00000074525.10
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr19_-_38031774 | 6.80 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr11_+_68936457 | 6.65 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr18_+_11766333 | 6.36 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr1_+_90842785 | 6.29 |
ENSMUST00000027528.7
|
Mlph
|
melanophilin |
| chr1_+_130659700 | 6.25 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr19_-_38032006 | 6.23 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr1_-_149836974 | 6.05 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr9_+_123902143 | 5.88 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr14_+_75373766 | 5.76 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr5_+_92957231 | 5.55 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr1_+_131671751 | 5.43 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr12_-_114252202 | 5.30 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr14_-_22039543 | 5.28 |
ENSMUST00000043409.9
|
Zfp503
|
zinc finger protein 503 |
| chr9_+_15150341 | 5.27 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr1_+_44590791 | 5.25 |
ENSMUST00000160854.8
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr18_+_44096300 | 5.23 |
ENSMUST00000069245.8
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr4_-_141325517 | 5.22 |
ENSMUST00000131317.8
ENSMUST00000006381.11 ENSMUST00000129602.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr11_+_116423266 | 5.15 |
ENSMUST00000106386.8
ENSMUST00000145737.8 ENSMUST00000155102.8 ENSMUST00000063446.13 |
Sphk1
|
sphingosine kinase 1 |
| chr19_-_5610628 | 5.10 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr5_-_142892457 | 5.10 |
ENSMUST00000167721.8
ENSMUST00000163829.2 ENSMUST00000100497.11 |
Actb
|
actin, beta |
| chr3_+_92272486 | 5.07 |
ENSMUST00000050397.2
|
Sprr2f
|
small proline-rich protein 2F |
| chr10_+_75407356 | 4.98 |
ENSMUST00000143226.8
ENSMUST00000124259.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr17_+_28426752 | 4.93 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr5_+_53195423 | 4.79 |
ENSMUST00000170523.8
|
Slc34a2
|
solute carrier family 34 (sodium phosphate), member 2 |
| chr14_-_21898992 | 4.79 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr7_-_44983080 | 4.79 |
ENSMUST00000211743.2
ENSMUST00000042194.10 |
Trpm4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr3_+_122522592 | 4.68 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr13_-_100922910 | 4.67 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr11_-_8614497 | 4.65 |
ENSMUST00000020695.13
|
Tns3
|
tensin 3 |
| chr19_-_24178000 | 4.61 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr11_-_99328969 | 4.59 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr8_+_70275079 | 4.42 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr2_+_3705824 | 4.39 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr7_+_130633776 | 4.32 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr18_-_64794338 | 4.25 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr2_-_148285450 | 4.22 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr2_+_16361582 | 4.17 |
ENSMUST00000114703.10
|
Plxdc2
|
plexin domain containing 2 |
| chr11_+_28803188 | 4.15 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chrX_-_36349055 | 4.09 |
ENSMUST00000115231.4
|
Rpl39
|
ribosomal protein L39 |
| chr2_+_84966569 | 4.09 |
ENSMUST00000057019.9
|
Aplnr
|
apelin receptor |
| chr5_-_142891686 | 3.94 |
ENSMUST00000106216.3
|
Actb
|
actin, beta |
| chr14_-_54651442 | 3.85 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr6_+_70675416 | 3.83 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr14_+_75373915 | 3.81 |
ENSMUST00000122840.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_-_167852538 | 3.76 |
ENSMUST00000099073.3
|
Ripor3
|
RIPOR family member 3 |
| chr6_+_67838100 | 3.73 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr1_-_133617824 | 3.72 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr12_-_114286421 | 3.72 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr6_-_52185674 | 3.66 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr15_-_97806142 | 3.58 |
ENSMUST00000023119.15
|
Vdr
|
vitamin D (1,25-dihydroxyvitamin D3) receptor |
| chr8_-_105991219 | 3.51 |
ENSMUST00000034359.10
|
Tradd
|
TNFRSF1A-associated via death domain |
| chr19_+_8906916 | 3.45 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr17_-_48758538 | 3.45 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2 |
| chr10_-_18421628 | 3.31 |
ENSMUST00000213153.2
|
Hebp2
|
heme binding protein 2 |
| chr9_-_99240571 | 3.27 |
ENSMUST00000042158.13
|
Esyt3
|
extended synaptotagmin-like protein 3 |
| chr19_-_40576897 | 3.26 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr8_+_3715747 | 3.24 |
ENSMUST00000014118.4
|
Mcemp1
|
mast cell expressed membrane protein 1 |
| chr2_-_164024649 | 3.22 |
ENSMUST00000045127.12
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr2_-_102016665 | 3.18 |
ENSMUST00000111222.2
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr3_-_90603013 | 3.15 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr11_-_8614667 | 3.12 |
ENSMUST00000239111.2
|
Tns3
|
tensin 3 |
| chr16_-_50252703 | 3.08 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr10_+_69048464 | 3.08 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr6_+_68916540 | 3.02 |
ENSMUST00000103339.2
|
Igkv13-84
|
immunoglobulin kappa chain variable 13-84 |
| chr5_+_114924106 | 3.01 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr11_+_48691175 | 2.96 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr14_+_79718604 | 2.94 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr10_+_13376745 | 2.91 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr19_-_40576782 | 2.89 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr11_+_23206001 | 2.88 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr1_+_61677977 | 2.87 |
ENSMUST00000075374.10
|
Pard3b
|
par-3 family cell polarity regulator beta |
| chr8_+_124059414 | 2.80 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2 |
| chr19_-_8690330 | 2.74 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr12_-_113700190 | 2.69 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr9_+_123901979 | 2.69 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr9_+_32283779 | 2.67 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr17_+_44445659 | 2.66 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr4_-_132149704 | 2.62 |
ENSMUST00000152271.8
ENSMUST00000084170.12 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr5_-_142891565 | 2.58 |
ENSMUST00000171419.8
|
Actb
|
actin, beta |
| chr15_-_79976016 | 2.56 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr6_+_22288220 | 2.54 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr17_+_32671689 | 2.54 |
ENSMUST00000237491.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr2_-_165997179 | 2.51 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr16_-_95928804 | 2.49 |
ENSMUST00000233292.2
ENSMUST00000050884.16 |
Hmgn1
|
high mobility group nucleosomal binding domain 1 |
| chr19_-_40576817 | 2.39 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr16_-_3725515 | 2.38 |
ENSMUST00000177221.2
ENSMUST00000177323.8 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr6_-_43643093 | 2.36 |
ENSMUST00000114644.8
ENSMUST00000067888.14 |
Tpk1
|
thiamine pyrophosphokinase |
| chr2_-_102016717 | 2.33 |
ENSMUST00000058790.12
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr1_+_34890912 | 2.33 |
ENSMUST00000152654.3
|
Plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr2_-_165997551 | 2.32 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr9_+_50768224 | 2.32 |
ENSMUST00000174628.8
ENSMUST00000034560.14 ENSMUST00000114437.9 ENSMUST00000175645.8 ENSMUST00000176349.8 ENSMUST00000176798.8 ENSMUST00000175640.8 |
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta |
| chr2_-_164024512 | 2.30 |
ENSMUST00000131974.2
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr5_+_33815466 | 2.30 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr17_+_28426831 | 2.29 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr12_-_54703281 | 2.28 |
ENSMUST00000056228.8
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr6_-_47790272 | 2.26 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr17_-_24746804 | 2.25 |
ENSMUST00000176353.8
ENSMUST00000176237.8 |
Traf7
|
TNF receptor-associated factor 7 |
| chr5_-_30278552 | 2.23 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr9_-_67739607 | 2.21 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr7_+_141027763 | 2.19 |
ENSMUST00000106003.2
|
Rplp2
|
ribosomal protein, large P2 |
| chr2_+_127696548 | 2.15 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr4_-_154110073 | 2.14 |
ENSMUST00000182191.8
ENSMUST00000146543.9 ENSMUST00000146426.2 |
Smim1
|
small integral membrane protein 1 |
| chr10_+_69048506 | 2.13 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr18_+_65564009 | 2.12 |
ENSMUST00000224056.3
ENSMUST00000049248.7 |
Malt1
|
MALT1 paracaspase |
| chr2_-_84605732 | 2.12 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr17_-_24746911 | 2.11 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7 |
| chr19_-_45738002 | 2.09 |
ENSMUST00000070215.8
|
Npm3
|
nucleoplasmin 3 |
| chr10_+_21870565 | 2.06 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_-_63663344 | 2.05 |
ENSMUST00000239480.2
ENSMUST00000062246.8 |
Tnfsf15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr7_+_43441315 | 2.05 |
ENSMUST00000005891.7
|
Klk9
|
kallikrein related-peptidase 9 |
| chr14_+_59716265 | 2.05 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chr16_-_16345197 | 2.03 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_+_110888313 | 2.02 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_-_74174524 | 1.99 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr5_+_135015046 | 1.99 |
ENSMUST00000094245.4
|
Cldn3
|
claudin 3 |
| chr6_-_34294377 | 1.99 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr2_-_84605764 | 1.97 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr17_-_71153283 | 1.94 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr10_+_79855454 | 1.93 |
ENSMUST00000043311.7
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr1_-_52230062 | 1.93 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr9_+_35334878 | 1.91 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr15_+_89206923 | 1.88 |
ENSMUST00000066991.7
|
Adm2
|
adrenomedullin 2 |
| chr3_-_58433313 | 1.87 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr17_-_34218301 | 1.87 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr3_+_79793237 | 1.86 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr11_+_69805005 | 1.85 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2 |
| chr17_+_35278011 | 1.79 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr12_+_24758968 | 1.79 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chrX_+_138464065 | 1.78 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr4_-_132149780 | 1.76 |
ENSMUST00000102568.10
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr14_-_34096574 | 1.76 |
ENSMUST00000023826.5
|
Sncg
|
synuclein, gamma |
| chr2_+_70339175 | 1.75 |
ENSMUST00000134607.8
|
Erich2
|
glutamate rich 2 |
| chr18_+_61688329 | 1.75 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr19_+_8907206 | 1.73 |
ENSMUST00000224272.2
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr6_-_142517340 | 1.73 |
ENSMUST00000203945.3
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr3_-_143908111 | 1.73 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr8_+_105991280 | 1.71 |
ENSMUST00000036221.12
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr1_-_173426640 | 1.68 |
ENSMUST00000150649.9
ENSMUST00000180215.2 ENSMUST00000097462.9 |
Ifi213
|
interferon activated gene 213 |
| chr1_-_85087567 | 1.68 |
ENSMUST00000161675.3
ENSMUST00000160792.8 |
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr7_+_141047416 | 1.68 |
ENSMUST00000209988.2
|
Cd151
|
CD151 antigen |
| chr11_-_45846291 | 1.67 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr2_+_70339157 | 1.66 |
ENSMUST00000100041.9
|
Erich2
|
glutamate rich 2 |
| chrX_+_7708295 | 1.60 |
ENSMUST00000115667.10
ENSMUST00000115668.10 ENSMUST00000115665.2 |
Otud5
|
OTU domain containing 5 |
| chr3_-_143908060 | 1.59 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr15_+_76929275 | 1.58 |
ENSMUST00000127957.8
ENSMUST00000129468.8 ENSMUST00000149569.9 ENSMUST00000152949.2 |
Apol6
|
apolipoprotein L 6 |
| chr3_+_63203516 | 1.54 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr1_-_194813843 | 1.53 |
ENSMUST00000075451.12
ENSMUST00000191775.2 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr5_-_34794185 | 1.53 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr5_+_33815910 | 1.53 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr1_+_16758629 | 1.51 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr4_-_154110324 | 1.51 |
ENSMUST00000130175.8
ENSMUST00000182151.8 |
Smim1
|
small integral membrane protein 1 |
| chr12_-_30423356 | 1.50 |
ENSMUST00000021004.14
|
Sntg2
|
syntrophin, gamma 2 |
| chr11_-_45845992 | 1.48 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr5_+_43819688 | 1.47 |
ENSMUST00000048150.15
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr12_+_105570350 | 1.47 |
ENSMUST00000041229.5
|
Bdkrb1
|
bradykinin receptor, beta 1 |
| chr14_+_30201569 | 1.45 |
ENSMUST00000022535.9
ENSMUST00000223658.2 |
Dcp1a
|
decapping mRNA 1A |
| chr5_+_107112186 | 1.45 |
ENSMUST00000117196.9
ENSMUST00000031221.12 ENSMUST00000076467.13 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr6_+_122967309 | 1.44 |
ENSMUST00000079379.3
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr14_+_26414422 | 1.43 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr7_+_101619019 | 1.43 |
ENSMUST00000084852.13
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chrX_+_7708034 | 1.40 |
ENSMUST00000033494.16
ENSMUST00000115666.8 |
Otud5
|
OTU domain containing 5 |
| chr18_+_61688378 | 1.36 |
ENSMUST00000165721.8
ENSMUST00000115246.9 ENSMUST00000166990.8 ENSMUST00000163205.8 ENSMUST00000170862.8 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr19_-_46917661 | 1.35 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr11_-_35871300 | 1.32 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr9_-_109063792 | 1.32 |
ENSMUST00000197329.2
ENSMUST00000079548.12 |
Fbxw20
|
F-box and WD-40 domain protein 20 |
| chr4_-_154110383 | 1.31 |
ENSMUST00000132541.8
ENSMUST00000143047.8 |
Smim1
|
small integral membrane protein 1 |
| chr4_+_107111106 | 1.30 |
ENSMUST00000046558.8
|
Hspb11
|
heat shock protein family B (small), member 11 |
| chr19_-_8906686 | 1.30 |
ENSMUST00000096242.5
|
Rom1
|
rod outer segment membrane protein 1 |
| chr4_+_107111083 | 1.30 |
ENSMUST00000106749.8
|
Hspb11
|
heat shock protein family B (small), member 11 |
| chr14_+_120543857 | 1.25 |
ENSMUST00000227594.2
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr7_+_113113292 | 1.24 |
ENSMUST00000122890.2
|
Far1
|
fatty acyl CoA reductase 1 |
| chr14_+_54312354 | 1.22 |
ENSMUST00000103679.3
|
Trdv4
|
T cell receptor delta variable 4 |
| chr2_+_89808124 | 1.19 |
ENSMUST00000061830.2
|
Olfr1260
|
olfactory receptor 1260 |
| chr5_-_100648487 | 1.15 |
ENSMUST00000239512.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr17_+_34812361 | 1.12 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr10_+_66932235 | 1.10 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chrX_+_163763588 | 1.10 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr15_-_77480311 | 1.09 |
ENSMUST00000089465.6
|
Apol10b
|
apolipoprotein L 10B |
| chr8_-_111573401 | 1.08 |
ENSMUST00000042012.7
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr1_+_173501215 | 1.06 |
ENSMUST00000085876.12
|
Ifi208
|
interferon activated gene 208 |
| chr16_-_32630847 | 1.04 |
ENSMUST00000179384.3
|
Smbd1
|
somatomedin B domain containing 1 |
| chr7_-_35096133 | 1.03 |
ENSMUST00000154597.2
ENSMUST00000032704.12 |
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chr11_-_102207516 | 1.03 |
ENSMUST00000107115.8
ENSMUST00000128016.2 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr11_-_102207486 | 1.01 |
ENSMUST00000146896.9
ENSMUST00000079589.11 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr11_+_75422516 | 1.01 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chrX_+_72683020 | 1.00 |
ENSMUST00000019701.9
|
Dusp9
|
dual specificity phosphatase 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.1 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 3.9 | 11.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 3.8 | 11.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 2.9 | 8.6 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 2.8 | 8.5 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 2.2 | 6.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.7 | 5.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.6 | 16.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.6 | 4.8 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 1.4 | 4.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.4 | 4.1 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 1.3 | 7.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.2 | 4.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.1 | 4.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.1 | 5.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.1 | 3.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.0 | 4.2 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.9 | 5.2 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.9 | 6.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.9 | 5.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.8 | 2.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.8 | 4.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.8 | 4.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.8 | 4.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.8 | 2.3 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.7 | 3.0 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.7 | 3.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.7 | 13.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.7 | 6.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.7 | 2.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.7 | 2.0 | GO:0090420 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.7 | 4.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 5.0 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 2.4 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.5 | 2.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.5 | 4.3 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.5 | 6.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 2.5 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 3.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.5 | 3.3 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.4 | 2.2 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.4 | 2.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.4 | 9.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.4 | 1.5 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.4 | 1.9 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.4 | 1.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.4 | 1.4 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.3 | 4.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 2.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 2.9 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 1.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 3.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 2.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.3 | 1.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 4.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.3 | 2.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 5.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 1.9 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.3 | 4.2 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.2 | 5.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 1.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 3.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 1.1 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.2 | 1.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.6 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 1.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 2.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 2.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 3.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.2 | 1.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 3.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.2 | 4.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 9.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 0.7 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.9 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.2 | 0.9 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.2 | 0.5 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 3.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.2 | 1.7 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 1.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 3.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 4.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 5.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 2.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 23.0 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.8 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 2.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 2.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 1.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.9 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 7.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 1.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.5 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 2.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 4.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 2.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 9.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.7 | GO:0010756 | regulation of plasminogen activation(GO:0010755) positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 5.5 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.1 | 1.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 2.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.7 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 | 0.2 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 2.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.9 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 0.8 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 2.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 4.6 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 3.4 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 9.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.2 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 1.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 2.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 2.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 7.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 2.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 3.7 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 4.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.7 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 1.7 | GO:0060348 | bone development(GO:0060348) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 35.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.2 | 4.7 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.1 | 6.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.0 | 5.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.0 | 3.0 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.4 | 3.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 2.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 21.2 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 6.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.3 | 4.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 1.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 1.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 1.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 2.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 7.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 3.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 4.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 0.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 6.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 3.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 2.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 4.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 6.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 8.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 5.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 10.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 6.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 13.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 7.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 9.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 14.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 6.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 8.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 4.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 6.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 14.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 5.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 8.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 2.5 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 7.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 2.9 | 8.6 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 2.8 | 8.5 | GO:0004349 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.9 | 19.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.5 | 35.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 1.4 | 4.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.2 | 3.6 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 1.1 | 3.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 1.0 | 4.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.0 | 2.9 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.0 | 4.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.8 | 4.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.7 | 4.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 6.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.6 | 5.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.5 | 4.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 4.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.5 | 4.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 1.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 2.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 2.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.5 | 6.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 4.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 5.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.5 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.4 | 3.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 3.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 2.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.3 | 7.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 1.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.9 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 6.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 5.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 2.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 2.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 0.9 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 2.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 4.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 7.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 5.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 2.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 3.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 4.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.2 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 4.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 0.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.8 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 2.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 2.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 1.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 18.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.2 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 4.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 22.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 3.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 12.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 3.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004080 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 10.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 5.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.5 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 7.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 6.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 11.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.5 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 6.5 | GO:0005543 | phospholipid binding(GO:0005543) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 12.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 11.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 5.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 6.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 5.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 16.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 6.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 13.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 3.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 9.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 16.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.0 | 8.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.4 | 4.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.3 | 6.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 11.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 5.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 2.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 4.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 5.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 6.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 6.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 4.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 5.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 6.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 9.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 4.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 8.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 9.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 10.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 7.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |