Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
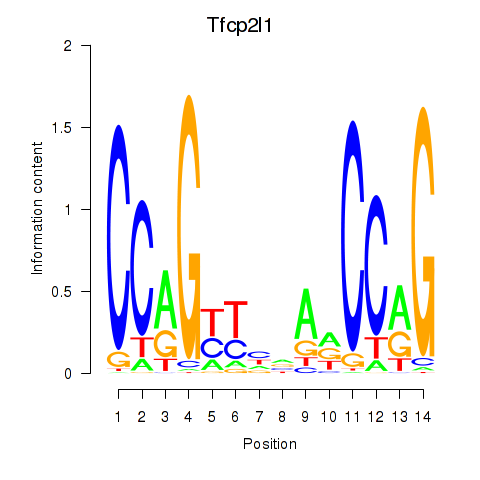
Results for Tfcp2l1
Z-value: 0.76
Transcription factors associated with Tfcp2l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2l1
|
ENSMUSG00000026380.11 | Tfcp2l1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2l1 | mm39_v1_chr1_+_118555668_118555686 | 0.43 | 1.9e-04 | Click! |
Activity profile of Tfcp2l1 motif
Sorted Z-values of Tfcp2l1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2l1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_121189091 | 7.00 |
ENSMUST00000000317.13
ENSMUST00000129130.3 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr11_-_116128903 | 4.79 |
ENSMUST00000037007.4
|
Evpl
|
envoplakin |
| chr17_-_26417982 | 4.42 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr1_+_130659700 | 4.28 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr2_-_163592127 | 4.06 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr7_-_140697719 | 3.49 |
ENSMUST00000067836.9
|
Ano9
|
anoctamin 9 |
| chr8_-_95405234 | 3.43 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr3_-_107425316 | 2.89 |
ENSMUST00000169449.8
ENSMUST00000029499.15 |
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr2_+_157266175 | 2.83 |
ENSMUST00000029175.14
ENSMUST00000092576.11 |
Src
|
Rous sarcoma oncogene |
| chr1_+_40720731 | 2.82 |
ENSMUST00000192345.2
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chrX_-_7185424 | 2.71 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr18_-_74197970 | 2.48 |
ENSMUST00000159162.2
ENSMUST00000091851.10 |
Mapk4
|
mitogen-activated protein kinase 4 |
| chr11_+_75422516 | 2.44 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr5_-_138262178 | 2.39 |
ENSMUST00000048421.14
|
Map11
|
microtubule associated protein 11 |
| chr3_+_138566249 | 2.32 |
ENSMUST00000121826.3
|
Tspan5
|
tetraspanin 5 |
| chr4_-_135300934 | 2.26 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr6_-_112923715 | 2.24 |
ENSMUST00000113169.9
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr11_+_42312150 | 2.16 |
ENSMUST00000192403.2
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr1_-_132635042 | 2.07 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr4_-_106585127 | 1.91 |
ENSMUST00000106770.8
ENSMUST00000145044.2 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr1_+_5153300 | 1.91 |
ENSMUST00000044369.13
ENSMUST00000194676.6 ENSMUST00000192029.6 ENSMUST00000192698.3 |
Atp6v1h
|
ATPase, H+ transporting, lysosomal V1 subunit H |
| chr9_+_32283779 | 1.85 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr1_-_132635078 | 1.82 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr11_-_105828624 | 1.76 |
ENSMUST00000183493.8
|
Cyb561
|
cytochrome b-561 |
| chr8_-_125161061 | 1.75 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr11_+_75422925 | 1.75 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr1_-_79838897 | 1.65 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chrX_-_7185529 | 1.64 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr2_-_91795910 | 1.64 |
ENSMUST00000239257.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chr17_-_36012932 | 1.62 |
ENSMUST00000166980.9
ENSMUST00000145900.8 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr6_-_28831746 | 1.59 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr10_-_81127334 | 1.51 |
ENSMUST00000219479.2
|
Tjp3
|
tight junction protein 3 |
| chr6_-_113911640 | 1.45 |
ENSMUST00000101044.9
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr2_-_26012751 | 1.45 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr3_-_107424637 | 1.44 |
ENSMUST00000166892.2
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr3_-_26207456 | 1.36 |
ENSMUST00000191835.2
|
Nlgn1
|
neuroligin 1 |
| chr18_+_36661198 | 1.35 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr10_-_81127057 | 1.35 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr3_-_59102517 | 1.35 |
ENSMUST00000200095.2
|
Gpr87
|
G protein-coupled receptor 87 |
| chr9_+_37450551 | 1.34 |
ENSMUST00000002008.7
ENSMUST00000215957.2 ENSMUST00000215271.2 |
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr14_+_25459630 | 1.32 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr2_-_120119544 | 1.30 |
ENSMUST00000094665.5
|
Pla2g4d
|
phospholipase A2, group IVD |
| chr3_+_55149947 | 1.26 |
ENSMUST00000167204.8
ENSMUST00000054237.14 |
Dclk1
|
doublecortin-like kinase 1 |
| chr16_-_76170714 | 1.25 |
ENSMUST00000231585.2
ENSMUST00000121927.8 |
Nrip1
|
nuclear receptor interacting protein 1 |
| chr19_+_6468761 | 1.24 |
ENSMUST00000113462.8
ENSMUST00000077182.13 ENSMUST00000236635.2 ENSMUST00000113461.8 |
Nrxn2
|
neurexin II |
| chr19_+_48194464 | 1.23 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr17_+_27171834 | 1.23 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr2_-_119060366 | 1.20 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr11_+_68979308 | 1.19 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr14_+_25459690 | 1.18 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr1_+_133109059 | 1.17 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr2_-_119060330 | 1.16 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr1_-_93088614 | 1.16 |
ENSMUST00000043718.12
|
Mab21l4
|
mab-21-like 4 |
| chr11_+_75422953 | 1.15 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr6_-_13838423 | 1.13 |
ENSMUST00000115492.2
|
Gpr85
|
G protein-coupled receptor 85 |
| chr8_+_57964921 | 1.13 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr9_+_43954681 | 1.08 |
ENSMUST00000114840.2
|
Thy1
|
thymus cell antigen 1, theta |
| chr9_+_32305259 | 1.06 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr3_-_94566107 | 1.06 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr8_-_123087485 | 1.02 |
ENSMUST00000044123.2
|
Trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr3_+_103482591 | 1.01 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chr2_+_79465761 | 1.01 |
ENSMUST00000111788.8
ENSMUST00000111784.9 |
Itprid2
|
ITPR interacting domain containing 2 |
| chr6_+_113448388 | 0.98 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr18_+_74198173 | 0.91 |
ENSMUST00000066583.2
|
Gm9925
|
predicted gene 9925 |
| chr7_+_105290259 | 0.90 |
ENSMUST00000209445.2
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr2_+_164611812 | 0.88 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr9_+_45049687 | 0.87 |
ENSMUST00000060125.7
|
Scn4b
|
sodium channel, type IV, beta |
| chr5_+_122988111 | 0.87 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chr3_+_106393348 | 0.86 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr2_+_79465696 | 0.84 |
ENSMUST00000111785.9
|
Itprid2
|
ITPR interacting domain containing 2 |
| chr16_-_21980200 | 0.83 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_-_78602313 | 0.82 |
ENSMUST00000229441.2
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr5_-_113229445 | 0.80 |
ENSMUST00000131708.2
ENSMUST00000117143.8 ENSMUST00000119627.8 |
Crybb3
|
crystallin, beta B3 |
| chr11_-_69493567 | 0.79 |
ENSMUST00000138694.2
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr17_-_36013189 | 0.79 |
ENSMUST00000135078.2
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr4_-_117744476 | 0.78 |
ENSMUST00000132073.2
ENSMUST00000150204.8 ENSMUST00000147845.2 ENSMUST00000036380.14 ENSMUST00000136596.2 |
Atp6v0b
|
ATPase, H+ transporting, lysosomal V0 subunit B |
| chr9_+_107765320 | 0.77 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr11_-_61065846 | 0.77 |
ENSMUST00000041683.9
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr6_-_83549399 | 0.76 |
ENSMUST00000206592.2
ENSMUST00000206400.2 |
Stambp
|
STAM binding protein |
| chr8_+_57964956 | 0.74 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr14_-_55950939 | 0.71 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr2_+_11783291 | 0.70 |
ENSMUST00000056108.12
|
Ankrd16
|
ankyrin repeat domain 16 |
| chrX_+_149829131 | 0.69 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr12_-_114487525 | 0.65 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr18_+_75953244 | 0.63 |
ENSMUST00000058997.15
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr12_+_17594795 | 0.63 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr7_-_100306160 | 0.61 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr3_+_103483397 | 0.61 |
ENSMUST00000183637.8
ENSMUST00000117221.9 ENSMUST00000118117.8 ENSMUST00000118563.3 |
Syt6
|
synaptotagmin VI |
| chr1_+_191873078 | 0.60 |
ENSMUST00000078470.12
ENSMUST00000110844.3 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr9_-_58065800 | 0.58 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr13_+_109769294 | 0.56 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr19_-_24454720 | 0.54 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr11_+_101215889 | 0.52 |
ENSMUST00000041095.14
ENSMUST00000107264.2 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr18_+_60880149 | 0.52 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr3_-_101743539 | 0.51 |
ENSMUST00000061831.11
|
Mab21l3
|
mab-21-like 3 |
| chr4_+_106768662 | 0.49 |
ENSMUST00000030367.15
ENSMUST00000149926.8 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr12_-_115944754 | 0.49 |
ENSMUST00000103551.2
|
Ighv1-84
|
immunoglobulin heavy variable 1-84 |
| chr1_-_143652711 | 0.47 |
ENSMUST00000159879.2
|
Ro60
|
Ro60, Y RNA binding protein |
| chr11_+_101221895 | 0.47 |
ENSMUST00000017316.7
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr6_+_91134358 | 0.45 |
ENSMUST00000155007.2
|
Hdac11
|
histone deacetylase 11 |
| chr17_-_33049169 | 0.44 |
ENSMUST00000201499.2
ENSMUST00000201876.4 ENSMUST00000202759.2 ENSMUST00000202988.4 |
Zfp799
|
zinc finger protein 799 |
| chr4_-_133695264 | 0.43 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr8_+_124100492 | 0.42 |
ENSMUST00000212571.2
ENSMUST00000212470.2 ENSMUST00000108840.4 ENSMUST00000057934.10 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr5_+_115568002 | 0.38 |
ENSMUST00000067168.9
|
Msi1
|
musashi RNA-binding protein 1 |
| chr14_-_55950545 | 0.38 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr1_+_171156942 | 0.36 |
ENSMUST00000111299.8
ENSMUST00000064950.11 |
Dedd
|
death effector domain-containing |
| chr10_+_80973787 | 0.34 |
ENSMUST00000117956.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr2_+_55327110 | 0.33 |
ENSMUST00000112633.3
ENSMUST00000112632.2 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr10_-_80382611 | 0.33 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr3_+_103482547 | 0.33 |
ENSMUST00000121834.8
|
Syt6
|
synaptotagmin VI |
| chr9_+_104930438 | 0.33 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr11_-_69768875 | 0.32 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr11_-_51497665 | 0.31 |
ENSMUST00000074669.10
ENSMUST00000101249.9 ENSMUST00000109103.4 |
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr3_-_62512561 | 0.30 |
ENSMUST00000058535.6
|
Gpr149
|
G protein-coupled receptor 149 |
| chr3_+_94391676 | 0.28 |
ENSMUST00000198384.3
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr8_-_72763462 | 0.28 |
ENSMUST00000003574.5
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr3_+_94391644 | 0.27 |
ENSMUST00000197677.5
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr14_+_34542053 | 0.26 |
ENSMUST00000043349.7
|
Grid1
|
glutamate receptor, ionotropic, delta 1 |
| chr1_+_171246593 | 0.25 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr15_-_78602971 | 0.23 |
ENSMUST00000088592.6
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr7_-_106390206 | 0.22 |
ENSMUST00000065024.2
|
Olfr699
|
olfactory receptor 699 |
| chr2_-_119916159 | 0.20 |
ENSMUST00000156159.4
|
Sptbn5
|
spectrin beta, non-erythrocytic 5 |
| chr18_+_37939442 | 0.18 |
ENSMUST00000076807.7
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr9_+_65278979 | 0.18 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr11_-_53508160 | 0.15 |
ENSMUST00000150568.8
|
Il4
|
interleukin 4 |
| chr17_+_35878332 | 0.15 |
ENSMUST00000044326.5
|
2300002M23Rik
|
RIKEN cDNA 2300002M23 gene |
| chr3_+_95071617 | 0.15 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr16_-_88548523 | 0.14 |
ENSMUST00000053149.4
|
Krtap13
|
keratin associated protein 13 |
| chr5_-_143278619 | 0.14 |
ENSMUST00000212355.2
|
Zfp853
|
zinc finger protein 853 |
| chr4_+_118818775 | 0.13 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr5_+_30805265 | 0.13 |
ENSMUST00000062962.12
|
Slc35f6
|
solute carrier family 35, member F6 |
| chr4_-_88562696 | 0.10 |
ENSMUST00000105149.3
|
Ifna13
|
interferon alpha 13 |
| chr15_-_101726600 | 0.09 |
ENSMUST00000023712.8
|
Krt2
|
keratin 2 |
| chr12_-_113860566 | 0.07 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr1_+_143653001 | 0.07 |
ENSMUST00000189936.7
ENSMUST00000018333.13 ENSMUST00000185493.7 |
Uchl5
|
ubiquitin carboxyl-terminal esterase L5 |
| chr13_-_115068626 | 0.06 |
ENSMUST00000056117.10
|
Itga2
|
integrin alpha 2 |
| chr1_+_86510614 | 0.04 |
ENSMUST00000121534.8
ENSMUST00000149542.2 |
Cops7b
|
COP9 signalosome subunit 7B |
| chr9_+_20914211 | 0.03 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chrX_+_48552803 | 0.03 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr9_-_42383494 | 0.02 |
ENSMUST00000128959.8
ENSMUST00000066148.12 ENSMUST00000138506.8 |
Tbcel
|
tubulin folding cofactor E-like |
| chr19_+_44748354 | 0.01 |
ENSMUST00000173346.4
|
Pax2
|
paired box 2 |
| chr7_-_96981221 | 0.00 |
ENSMUST00000139582.9
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr13_-_99584091 | 0.00 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0006154 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 1.2 | 3.5 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.9 | 2.8 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.5 | 1.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.5 | 4.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.5 | 1.4 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.5 | 1.4 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.4 | 2.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 3.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 2.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 3.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 2.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 2.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 0.9 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 1.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 2.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.2 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.2 | 0.5 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.2 | 0.8 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 2.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.6 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 4.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 4.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.3 | GO:0042376 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 2.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 2.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 1.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 4.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 5.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.6 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 1.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.2 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 1.2 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 2.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 7.0 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 1.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 1.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.2 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.9 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.0 | 0.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 3.5 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 1.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 0.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 3.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 4.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 7.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 4.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 1.8 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.4 | 4.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 2.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 3.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 1.0 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 4.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 2.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 2.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 2.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 3.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 2.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 4.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 1.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 4.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 3.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 4.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 5.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0016775 | phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 2.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.6 | GO:0004143 | lipid kinase activity(GO:0001727) diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.1 | 2.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 1.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 2.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 2.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 8.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 5.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 5.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 3.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 4.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |