Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
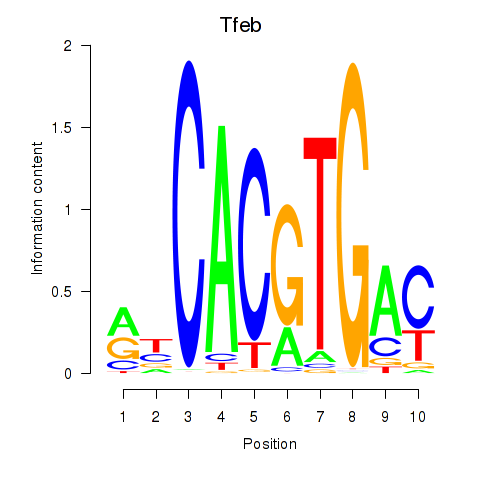
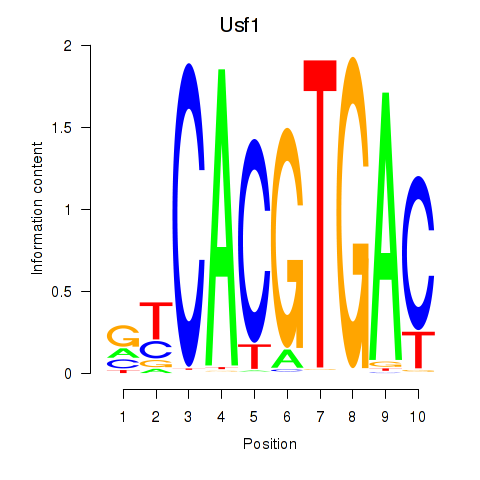
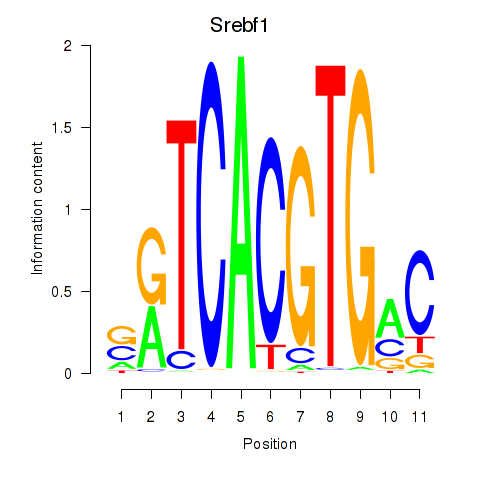
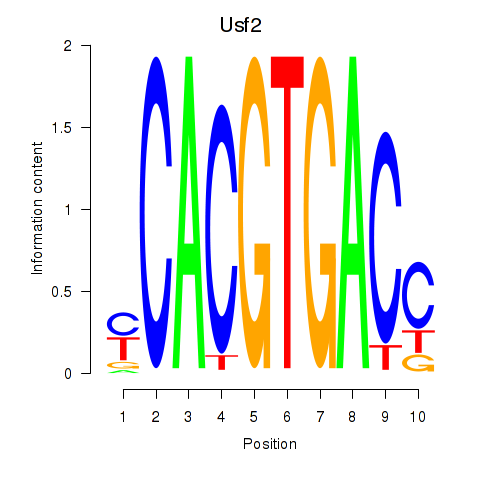
Results for Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Z-value: 2.32
Transcription factors associated with Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfeb
|
ENSMUSG00000023990.19 | Tfeb |
|
Usf1
|
ENSMUSG00000026641.14 | Usf1 |
|
Srebf1
|
ENSMUSG00000020538.16 | Srebf1 |
|
Usf2
|
ENSMUSG00000058239.14 | Usf2 |
|
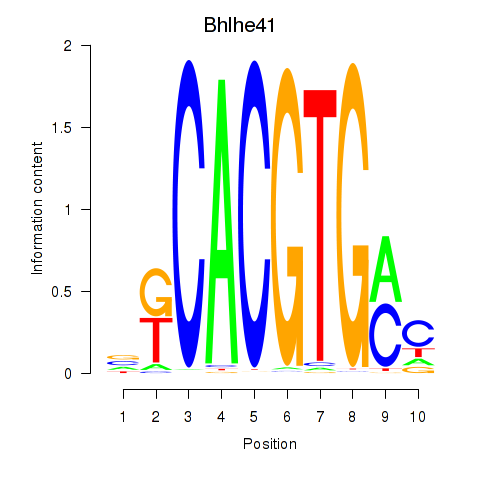
Bhlhe41
|
ENSMUSG00000030256.12 | Bhlhe41 |
|
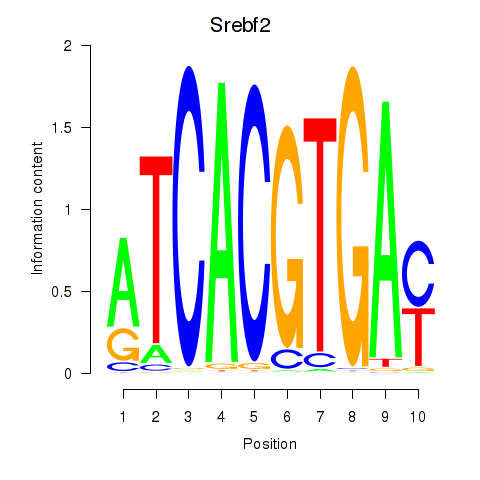
Srebf2
|
ENSMUSG00000022463.9 | Srebf2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srebf2 | mm39_v1_chr15_+_82031382_82031476 | 0.71 | 2.9e-12 | Click! |
| Usf1 | mm39_v1_chr1_+_171238873_171238906 | 0.63 | 2.1e-09 | Click! |
| Bhlhe41 | mm39_v1_chr6_-_145811274_145811284 | 0.60 | 2.4e-08 | Click! |
| Usf2 | mm39_v1_chr7_-_30656167_30656228 | 0.50 | 9.2e-06 | Click! |
| Srebf1 | mm39_v1_chr11_-_60111391_60111458 | -0.22 | 5.8e-02 | Click! |
| Tfeb | mm39_v1_chr17_+_48047955_48048033 | 0.19 | 1.0e-01 | Click! |
Activity profile of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 motif
Sorted Z-values of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.7 | 53.0 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 10.1 | 50.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 5.3 | 42.0 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 4.9 | 19.8 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 4.7 | 51.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 4.7 | 23.6 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 4.7 | 28.2 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 4.6 | 32.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 3.9 | 15.6 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 3.5 | 21.0 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 3.5 | 13.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 3.5 | 20.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 3.4 | 20.6 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 3.3 | 13.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 3.1 | 9.4 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 3.1 | 15.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 3.0 | 15.0 | GO:0015808 | L-alanine transport(GO:0015808) |
| 2.8 | 8.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.8 | 8.5 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 2.8 | 33.9 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 2.8 | 28.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 2.8 | 13.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 2.8 | 13.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.8 | 11.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 2.7 | 8.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 2.7 | 18.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 2.6 | 12.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 2.6 | 64.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 2.5 | 7.6 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 2.5 | 170.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 2.5 | 20.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 2.5 | 10.0 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 2.5 | 9.9 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 2.5 | 39.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 2.3 | 6.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 2.2 | 8.9 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 2.1 | 15.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 2.1 | 12.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 2.1 | 12.5 | GO:0032439 | endosome localization(GO:0032439) |
| 2.0 | 19.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 2.0 | 11.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 2.0 | 9.8 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 1.9 | 9.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 1.9 | 18.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.9 | 5.6 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 1.9 | 7.4 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.9 | 5.6 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 1.8 | 11.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 1.8 | 10.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.7 | 6.8 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 1.7 | 13.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 1.6 | 4.9 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 1.6 | 14.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.6 | 6.4 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 1.6 | 46.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 1.6 | 4.7 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 1.6 | 14.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.6 | 9.4 | GO:0042117 | monocyte activation(GO:0042117) |
| 1.6 | 17.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.4 | 10.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 1.4 | 4.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.3 | 2.7 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.3 | 5.3 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 1.3 | 10.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.3 | 7.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.2 | 6.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 1.2 | 4.9 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.2 | 4.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.2 | 2.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 1.2 | 4.9 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 1.2 | 17.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 1.2 | 8.5 | GO:0010288 | response to lead ion(GO:0010288) |
| 1.2 | 3.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.2 | 3.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 1.1 | 3.3 | GO:0090650 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.1 | 6.7 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 1.1 | 15.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.1 | 3.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.1 | 7.6 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 1.1 | 8.7 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.0 | 6.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.0 | 12.4 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 1.0 | 7.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.0 | 7.9 | GO:0090306 | polar body extrusion after meiotic divisions(GO:0040038) spindle assembly involved in meiosis(GO:0090306) |
| 1.0 | 6.9 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 1.0 | 2.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.0 | 2.9 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 1.0 | 6.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 1.0 | 13.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 1.0 | 6.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.0 | 2.9 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.9 | 3.7 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.9 | 2.8 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.9 | 13.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.9 | 5.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.9 | 8.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.9 | 5.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.9 | 8.9 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.9 | 22.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.9 | 4.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.8 | 7.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.8 | 6.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.8 | 7.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.8 | 10.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.8 | 8.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.8 | 1.5 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.8 | 20.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.8 | 3.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.8 | 4.5 | GO:0030421 | defecation(GO:0030421) |
| 0.7 | 9.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 55.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.7 | 4.2 | GO:0045876 | DNA replication, removal of RNA primer(GO:0043137) positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.7 | 4.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.7 | 7.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.7 | 4.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.7 | 10.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.7 | 7.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.7 | 11.8 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.7 | 11.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.7 | 10.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.7 | 3.3 | GO:0061198 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.6 | 11.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.6 | 14.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.6 | 12.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.6 | 2.5 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.6 | 8.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.6 | 1.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 7.2 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.6 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 9.5 | GO:0043084 | penile erection(GO:0043084) |
| 0.6 | 2.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.6 | 5.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.6 | 11.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 1.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.6 | 13.6 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.6 | 1.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.6 | 5.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.6 | 12.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.5 | 2.2 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.5 | 8.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.5 | 1.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.5 | 3.6 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.5 | 4.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.5 | 2.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.5 | 4.0 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.5 | 2.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.5 | 5.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.5 | 2.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 5.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.5 | 3.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 8.6 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.5 | 2.9 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.5 | 0.5 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.5 | 6.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 1.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.5 | 2.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 3.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 5.1 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.5 | 9.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.5 | 0.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 4.6 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.5 | 1.4 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.4 | 7.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 | 1.8 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.4 | 18.3 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.4 | 12.9 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.4 | 2.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 1.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.4 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.4 | 1.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.3 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.4 | 3.0 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.4 | 5.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 2.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 | 1.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.4 | 3.6 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.4 | 0.4 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.4 | 1.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.4 | 0.8 | GO:0019858 | MAPK import into nucleus(GO:0000189) cytosine metabolic process(GO:0019858) |
| 0.4 | 3.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.4 | 1.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.4 | 1.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 5.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.4 | 0.8 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.4 | 2.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.4 | 1.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 6.7 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.4 | 1.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 0.7 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.4 | 1.5 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.4 | 1.5 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.4 | 3.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 2.8 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.4 | 4.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 0.7 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 1.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 5.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 0.6 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 0.6 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.3 | 0.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.3 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 | 2.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.3 | 3.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.3 | 3.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.6 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.3 | 9.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.3 | 5.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 0.9 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 2.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.3 | 4.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 6.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.3 | 12.9 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.3 | 1.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.3 | 0.9 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.3 | 1.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 2.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 5.0 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.3 | 2.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 11.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.3 | 4.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.3 | 29.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.3 | 5.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 2.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 10.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.3 | 2.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 2.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 1.4 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.3 | 4.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 6.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.3 | 11.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.3 | 5.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.3 | 3.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 | 9.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 1.3 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 | 2.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 1.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 8.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 1.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 10.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 1.6 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 9.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.3 | 4.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 1.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 1.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 12.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.2 | 2.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 6.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 0.7 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 1.5 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 3.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 6.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.2 | 0.9 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 0.7 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.2 | 0.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 5.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 4.0 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.9 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 3.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 5.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.9 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 0.7 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 2.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 2.2 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.2 | 1.1 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 0.7 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 | 2.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 2.2 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.2 | 0.9 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 1.5 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 | 2.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 2.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.2 | 2.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 1.9 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.2 | 2.3 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.2 | 3.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 3.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 1.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 5.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 19.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.2 | 4.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 1.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 1.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 12.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 5.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 1.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 0.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 1.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 10.7 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.2 | 12.9 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.2 | 1.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 16.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 19.5 | GO:0043473 | pigmentation(GO:0043473) |
| 0.2 | 5.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.2 | 1.9 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 2.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 5.0 | GO:1903205 | regulation of hydrogen peroxide-induced cell death(GO:1903205) |
| 0.2 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 0.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.7 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 0.9 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.2 | 3.3 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.2 | 1.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 1.4 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 | 10.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 6.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 5.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.5 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.2 | 0.5 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 18.7 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.2 | 0.7 | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) |
| 0.2 | 8.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 1.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 1.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.2 | 3.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 1.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.2 | 2.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 0.9 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) phenotypic switching(GO:0036166) cellular response to cocaine(GO:0071314) |
| 0.1 | 2.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 1.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 4.7 | GO:0034728 | nucleosome organization(GO:0034728) |
| 0.1 | 0.4 | GO:1990414 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 2.0 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.6 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 1.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 1.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 3.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 1.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 2.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 8.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 3.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 7.3 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 1.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 | 0.6 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.5 | GO:0071468 | cellular response to acidic pH(GO:0071468) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 2.8 | GO:0043094 | cellular metabolic compound salvage(GO:0043094) |
| 0.1 | 8.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 3.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.7 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.1 | 1.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 8.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.3 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 0.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 6.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.5 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 2.0 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 4.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 2.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 3.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.8 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 1.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.7 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.3 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.1 | 1.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 1.8 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.1 | 1.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 3.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.5 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 1.6 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.1 | 4.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.5 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 3.0 | GO:1904591 | positive regulation of protein import(GO:1904591) |
| 0.1 | 1.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 6.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 16.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 4.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 3.1 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.9 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 0.3 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.1 | 2.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 2.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 2.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.6 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 2.0 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 2.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 14.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 0.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 4.8 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.1 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 4.9 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 5.2 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 1.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.3 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 0.5 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 0.9 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.2 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 2.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.7 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.8 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.2 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 0.2 | GO:0061188 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.2 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.1 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 2.1 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 1.6 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 1.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 1.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 1.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.9 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 6.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 3.0 | GO:0051283 | release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 2.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.5 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.2 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.9 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 4.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 10.2 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.0 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.4 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 1.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 13.8 | GO:0016568 | chromatin modification(GO:0016568) |
| 0.0 | 1.9 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 1.5 | GO:1904029 | regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 4.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 5.3 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.6 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.1 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 1.2 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 2.2 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0048010 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.0 | 0.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 1.0 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 3.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.7 | GO:0007030 | Golgi organization(GO:0007030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.7 | 53.0 | GO:0097144 | BAX complex(GO:0097144) |
| 11.2 | 56.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 7.5 | 29.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 6.7 | 20.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 6.6 | 79.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 6.4 | 32.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 6.2 | 18.7 | GO:1990879 | CST complex(GO:1990879) |
| 5.1 | 41.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 4.1 | 32.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 3.7 | 18.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 3.1 | 9.4 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 3.0 | 15.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 2.9 | 11.5 | GO:0019034 | viral replication complex(GO:0019034) |
| 2.8 | 19.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 2.8 | 27.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 2.7 | 10.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 2.4 | 14.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.9 | 3.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.8 | 5.4 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 1.8 | 5.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 1.8 | 19.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.7 | 14.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.7 | 12.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.6 | 20.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.5 | 6.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.5 | 12.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.5 | 9.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.5 | 4.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.5 | 22.7 | GO:0071203 | WASH complex(GO:0071203) |
| 1.5 | 7.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.5 | 14.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.4 | 8.6 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 1.4 | 9.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.3 | 33.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.3 | 12.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 1.2 | 8.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.2 | 7.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.1 | 9.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 1.1 | 6.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.1 | 6.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.1 | 11.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 1.1 | 84.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 1.1 | 8.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.1 | 14.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 1.0 | 11.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 1.0 | 6.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.0 | 8.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.0 | 4.9 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.9 | 9.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.9 | 3.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.9 | 21.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.9 | 9.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.8 | 3.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 13.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.8 | 12.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 16.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.8 | 7.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.7 | 5.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.7 | 3.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.7 | 6.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.7 | 7.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.7 | 9.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.7 | 12.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.6 | 9.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.6 | 3.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.6 | 1.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 4.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 11.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.5 | 3.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 7.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.5 | 3.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.5 | 120.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.5 | 14.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.5 | 2.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 1.4 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.5 | 1.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.5 | 3.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 1.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 6.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.4 | 2.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 4.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 4.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 5.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 0.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 5.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 11.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 1.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.4 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.4 | 16.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 2.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 3.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 8.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 6.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 10.4 | GO:0098793 | presynapse(GO:0098793) |
| 0.3 | 0.9 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 11.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 88.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 3.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 1.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 6.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 1.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 4.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 2.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 1.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 13.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 8.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 2.0 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 2.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 8.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 7.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 11.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 4.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 7.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 17.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 8.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 2.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 1.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 4.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.6 | GO:0005712 | chiasma(GO:0005712) |
| 0.2 | 36.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 3.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 3.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 1.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 5.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 2.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 5.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 10.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 5.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 2.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 10.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 2.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.7 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 2.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 22.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.2 | 15.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 159.6 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.2 | 11.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 0.5 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.2 | 6.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 2.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 13.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 1.7 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 4.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 15.3 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 0.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 18.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 7.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.6 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.1 | 2.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 12.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 11.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 30.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.6 | GO:0030135 | coated vesicle(GO:0030135) |
| 0.1 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 40.9 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 27.3 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 0.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 3.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 7.6 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 5.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 14.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 20.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 15.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 21.9 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.1 | 22.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 2.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 1.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 3.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 3.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 33.5 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 1.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 3.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.7 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 2.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.4 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 4.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 41.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 17.8 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 0.8 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 4.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 10.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 22.5 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.8 | 50.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 6.3 | 31.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 6.2 | 18.7 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 5.3 | 53.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 5.0 | 15.0 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 3.9 | 11.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 3.7 | 11.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 3.4 | 23.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 3.3 | 49.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 3.0 | 98.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 2.9 | 14.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 2.7 | 13.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 2.6 | 7.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 2.5 | 14.8 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 2.4 | 4.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 2.4 | 23.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 2.3 | 7.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 2.3 | 6.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 2.2 | 49.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 2.2 | 2.2 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 2.1 | 8.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.1 | 14.8 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 2.1 | 8.5 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 2.1 | 14.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 2.0 | 10.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 2.0 | 17.9 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 2.0 | 13.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.9 | 7.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.8 | 10.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.7 | 5.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.7 | 44.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 1.7 | 6.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.6 | 50.6 | GO:0005123 | death receptor binding(GO:0005123) |
| 1.6 | 4.9 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.6 | 11.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.4 | 11.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.4 | 7.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.4 | 8.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.4 | 5.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 1.4 | 161.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 1.3 | 8.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.3 | 4.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.2 | 5.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 1.2 | 13.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 1.2 | 4.9 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 1.2 | 13.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.2 | 3.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.2 | 6.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.2 | 17.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.1 | 15.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.1 | 13.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 1.0 | 11.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.0 | 6.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.0 | 2.9 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.0 | 2.9 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.9 | 1.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.9 | 9.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.9 | 3.6 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.9 | 9.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.9 | 12.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 52.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.9 | 2.7 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.8 | 4.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.8 | 2.5 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.8 | 6.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 11.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.8 | 3.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.8 | 3.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.8 | 23.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.7 | 3.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.7 | 9.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.7 | 3.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.7 | 6.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.7 | 2.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.7 | 12.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.7 | 24.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.7 | 2.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.7 | 33.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.7 | 6.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.7 | 18.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.7 | 2.0 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.6 | 13.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.6 | 3.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.6 | 11.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 2.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 12.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 2.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.6 | 3.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 10.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 13.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 10.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 18.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.6 | 2.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.6 | 2.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.6 | 3.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.6 | 2.8 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.6 | 3.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.5 | 2.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.5 | 5.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 8.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.5 | 2.1 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.5 | 6.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 16.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 9.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 5.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 5.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 5.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 2.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 1.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.5 | 1.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.5 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.5 | 1.5 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.5 | 2.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.5 | 4.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 1.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.5 | 5.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 3.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 11.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 15.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.4 | 2.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 1.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 5.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 1.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 7.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.4 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.4 | 8.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.4 | 3.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 6.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 1.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 5.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 5.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 3.9 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.4 | 2.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 6.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 1.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 2.7 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 4.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 15.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 4.9 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.4 | 4.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 3.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.4 | 1.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 4.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 25.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 7.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.4 | 2.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 2.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 4.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 2.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 7.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 4.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 1.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 1.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 30.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.3 | 6.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 1.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.3 | 24.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.3 | 0.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 0.9 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.3 | 1.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 10.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 1.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 13.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 2.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 4.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 1.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 2.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 4.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 28.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.3 | 28.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 13.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 1.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 3.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 86.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 18.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 4.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 102.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.3 | 1.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.0 | GO:0051381 | histamine binding(GO:0051381) |
| 0.2 | 2.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 9.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 6.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.0 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 7.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 3.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 2.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 2.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 4.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 12.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 1.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 3.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 12.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 1.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 5.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 4.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 5.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 1.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.6 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.2 | 2.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 13.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 9.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 18.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 1.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 14.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 3.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 6.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 2.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 1.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 8.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.2 | 1.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.7 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 0.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 1.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 3.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 9.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.7 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 6.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 7.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 2.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 2.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 6.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.4 | GO:0061749 | forked DNA-dependent helicase activity(GO:0061749) |
| 0.1 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 16.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 4.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 9.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 6.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.5 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 1.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 23.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 8.6 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 4.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 6.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 2.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.5 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 0.3 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.1 | 7.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 25.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 2.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 8.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 4.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 2.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 1.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 3.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 13.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 3.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 10.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 6.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 28.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 4.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 2.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 3.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 1.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 3.4 | GO:0005253 | anion channel activity(GO:0005253) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.1 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 4.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 5.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 5.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 6.9 | GO:0032403 | protein complex binding(GO:0032403) |
| 0.0 | 1.6 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 53.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.5 | 26.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.2 | 45.1 | PID ATM PATHWAY | ATM pathway |
| 0.8 | 14.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 8.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.6 | 13.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.5 | 15.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.5 | 57.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.5 | 10.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.5 | 15.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 19.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.4 | 9.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.4 | 10.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.4 | 18.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 8.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 7.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 6.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 13.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 14.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 9.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 7.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 18.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 4.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 4.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 5.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 9.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 11.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 3.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 5.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 6.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 2.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 11.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 14.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 5.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 6.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 10.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 4.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 13.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 15.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 6.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 93.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 1.0 | 55.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.9 | 43.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.9 | 39.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.8 | 13.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.7 | 16.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.7 | 26.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.6 | 9.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.6 | 18.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.6 | 8.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.6 | 8.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.6 | 17.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 3.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.5 | 19.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.5 | 18.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.5 | 10.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 5.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 4.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 6.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.4 | 13.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.4 | 5.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 4.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 5.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.4 | 9.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 6.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 22.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 1.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.3 | 8.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 6.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 4.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 6.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 17.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 3.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.3 | 24.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 4.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 3.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.3 | 24.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 1.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.9 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.2 | 6.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 7.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 4.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 3.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 25.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 2.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 4.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 1.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 15.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 3.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 16.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 21.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 2.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 7.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.9 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 25.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 14.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 6.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 8.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 2.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 7.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 5.5 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 13.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 5.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 6.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 15.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 3.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |