Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
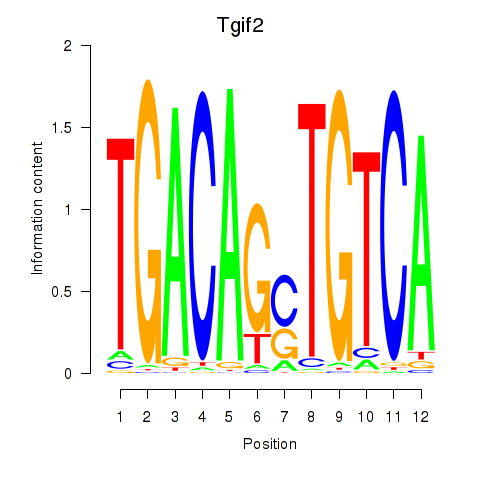
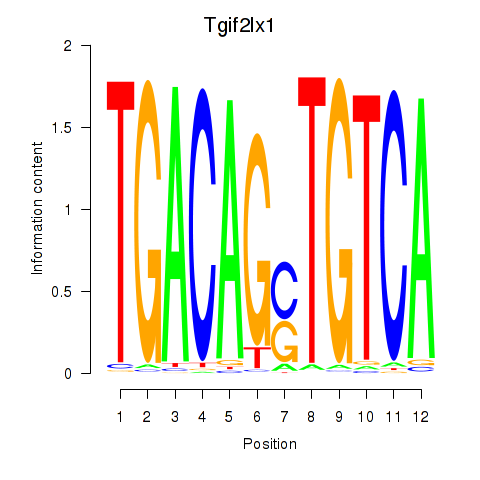
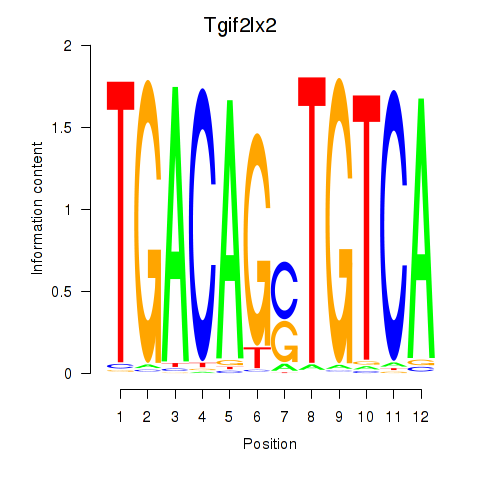
Results for Tgif2_Tgif2lx1_Tgif2lx2
Z-value: 1.18
Transcription factors associated with Tgif2_Tgif2lx1_Tgif2lx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tgif2
|
ENSMUSG00000062175.14 | Tgif2 |
|
Tgif2lx1
|
ENSMUSG00000100133.2 | Tgif2lx1 |
|
Tgif2lx2
|
ENSMUSG00000100194.2 | Tgif2lx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tgif2lx1 | mm39_v1_chrX_-_117390434_117390434 | -0.23 | 5.7e-02 | Click! |
| Tgif2lx2 | mm39_v1_chrX_+_117336912_117336928 | -0.22 | 6.4e-02 | Click! |
| Tgif2 | mm39_v1_chr2_+_156681927_156681952 | -0.16 | 1.9e-01 | Click! |
Activity profile of Tgif2_Tgif2lx1_Tgif2lx2 motif
Sorted Z-values of Tgif2_Tgif2lx1_Tgif2lx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tgif2_Tgif2lx1_Tgif2lx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_19144950 | 21.49 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr6_+_112436466 | 13.44 |
ENSMUST00000075477.8
|
Cav3
|
caveolin 3 |
| chr13_+_54849268 | 9.43 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr4_+_42950367 | 6.82 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr11_+_115768323 | 6.64 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chrX_+_93410718 | 6.17 |
ENSMUST00000113898.8
|
Apoo
|
apolipoprotein O |
| chrX_+_93410754 | 6.08 |
ENSMUST00000113897.9
ENSMUST00000113896.8 ENSMUST00000113895.2 |
Apoo
|
apolipoprotein O |
| chr2_-_84652890 | 5.96 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr3_+_107784543 | 5.87 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr13_-_63006176 | 4.80 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr16_-_88424546 | 4.62 |
ENSMUST00000052337.4
|
2310079G19Rik
|
RIKEN cDNA 2310079G19 gene |
| chr2_-_17395765 | 4.18 |
ENSMUST00000177966.2
|
Nebl
|
nebulette |
| chr16_+_33614378 | 3.90 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr6_-_68609426 | 3.44 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr1_+_36346824 | 3.29 |
ENSMUST00000142319.8
ENSMUST00000097778.9 ENSMUST00000115031.8 ENSMUST00000115032.8 ENSMUST00000137906.2 ENSMUST00000115029.2 |
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr4_+_148727774 | 3.28 |
ENSMUST00000105698.3
|
Gm572
|
predicted gene 572 |
| chr11_+_98239230 | 3.27 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr8_-_123187406 | 3.21 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr2_+_110427643 | 3.17 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr8_-_71292295 | 3.11 |
ENSMUST00000212405.2
ENSMUST00000002989.11 |
Arrdc2
|
arrestin domain containing 2 |
| chr11_+_101221895 | 3.08 |
ENSMUST00000017316.7
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr7_-_44320244 | 2.95 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr11_+_76792977 | 2.88 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr6_-_88851579 | 2.80 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr6_-_88852017 | 2.76 |
ENSMUST00000145944.3
|
Podxl2
|
podocalyxin-like 2 |
| chr6_+_112250719 | 2.38 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr1_-_52766615 | 2.37 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr4_-_87724533 | 2.31 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_-_68681962 | 2.16 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr1_+_135980488 | 2.15 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr6_-_69792108 | 2.12 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chrX_-_101687813 | 2.12 |
ENSMUST00000052012.14
ENSMUST00000043596.12 ENSMUST00000119229.8 ENSMUST00000122022.8 ENSMUST00000120270.8 ENSMUST00000113611.3 |
Phka1
|
phosphorylase kinase alpha 1 |
| chr17_+_34573760 | 2.09 |
ENSMUST00000178562.2
ENSMUST00000025198.15 |
Btnl2
|
butyrophilin-like 2 |
| chr1_+_135980639 | 2.05 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr1_+_135980508 | 2.03 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr6_-_70292451 | 2.01 |
ENSMUST00000103387.3
|
Igkv8-21
|
immunoglobulin kappa variable 8-21 |
| chr11_+_87000032 | 1.99 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_67939702 | 1.98 |
ENSMUST00000097290.4
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr17_+_44445659 | 1.98 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr2_+_157298836 | 1.94 |
ENSMUST00000109529.2
|
Src
|
Rous sarcoma oncogene |
| chr11_+_103540391 | 1.93 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr2_+_26209755 | 1.89 |
ENSMUST00000066889.13
|
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr1_-_106980033 | 1.89 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr10_+_5589210 | 1.89 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr9_+_48273333 | 1.86 |
ENSMUST00000093853.5
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr4_-_116024788 | 1.78 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr16_+_6887689 | 1.73 |
ENSMUST00000229741.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_29309942 | 1.68 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr4_-_87724512 | 1.67 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr19_+_42024439 | 1.64 |
ENSMUST00000238137.2
ENSMUST00000026172.3 |
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr9_-_71499547 | 1.64 |
ENSMUST00000169573.8
|
Myzap
|
myocardial zonula adherens protein |
| chr3_-_63391300 | 1.55 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr8_+_3705760 | 1.55 |
ENSMUST00000169234.3
ENSMUST00000012849.15 |
Retn
|
resistin |
| chr5_-_134581235 | 1.55 |
ENSMUST00000036999.10
ENSMUST00000100647.7 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr6_-_69835868 | 1.54 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr19_-_12093187 | 1.53 |
ENSMUST00000208391.3
ENSMUST00000214103.2 |
Olfr1428
|
olfactory receptor 1428 |
| chr7_-_142223662 | 1.53 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr4_-_147989137 | 1.52 |
ENSMUST00000105716.9
ENSMUST00000105715.8 ENSMUST00000105714.8 ENSMUST00000030884.10 |
Mfn2
|
mitofusin 2 |
| chr9_+_46151994 | 1.52 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr17_+_6320731 | 1.51 |
ENSMUST00000088940.6
|
Tmem181a
|
transmembrane protein 181A |
| chr9_+_44045859 | 1.51 |
ENSMUST00000034650.15
ENSMUST00000098852.3 ENSMUST00000216002.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr1_+_128079543 | 1.49 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr7_-_83384711 | 1.48 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr10_-_87982732 | 1.48 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr7_+_106692427 | 1.47 |
ENSMUST00000210568.4
|
Olfr17
|
olfactory receptor 17 |
| chr4_+_123681649 | 1.46 |
ENSMUST00000053202.12
|
Rhbdl2
|
rhomboid like 2 |
| chr3_+_134534754 | 1.41 |
ENSMUST00000029822.6
|
Tacr3
|
tachykinin receptor 3 |
| chr6_-_69741999 | 1.39 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr3_+_96577447 | 1.39 |
ENSMUST00000048427.9
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr11_+_31950452 | 1.37 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr9_-_71499628 | 1.37 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr6_-_33037191 | 1.31 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr6_-_33037107 | 1.25 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr5_-_115257336 | 1.24 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr8_+_123338357 | 1.22 |
ENSMUST00000015157.10
|
Trappc2l
|
trafficking protein particle complex 2-like |
| chr6_-_99643723 | 1.18 |
ENSMUST00000032151.3
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3 |
| chr7_+_48438751 | 1.16 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr2_-_7400780 | 1.13 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr19_-_33369655 | 1.12 |
ENSMUST00000163093.2
|
Rnls
|
renalase, FAD-dependent amine oxidase |
| chr1_+_118409769 | 1.06 |
ENSMUST00000191823.6
|
Clasp1
|
CLIP associating protein 1 |
| chr5_-_100307130 | 1.05 |
ENSMUST00000139520.3
|
Tmem150c
|
transmembrane protein 150C |
| chr15_-_33687986 | 1.01 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr6_-_52190299 | 1.00 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr11_-_86999481 | 0.98 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr2_-_7400690 | 0.98 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr15_+_79784365 | 0.98 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr3_-_146476331 | 0.97 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr3_-_146475974 | 0.95 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr15_-_85695855 | 0.95 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr7_-_107357104 | 0.94 |
ENSMUST00000208217.2
ENSMUST00000052438.8 |
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr16_-_92156312 | 0.94 |
ENSMUST00000051705.7
|
Kcne1
|
potassium voltage-gated channel, Isk-related subfamily, member 1 |
| chr15_-_76084776 | 0.93 |
ENSMUST00000169108.8
ENSMUST00000170728.8 |
Plec
|
plectin |
| chr1_-_63253702 | 0.90 |
ENSMUST00000050536.14
|
Gpr1
|
G protein-coupled receptor 1 |
| chr5_-_103359117 | 0.90 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr7_+_130972867 | 0.89 |
ENSMUST00000075610.13
|
Pstk
|
phosphoseryl-tRNA kinase |
| chr11_-_5691117 | 0.89 |
ENSMUST00000140922.2
ENSMUST00000093362.12 |
Urgcp
|
upregulator of cell proliferation |
| chr10_+_81395242 | 0.87 |
ENSMUST00000002518.9
|
Tle5
|
TLE family member 5, transcriptional modulator |
| chr4_-_123611974 | 0.87 |
ENSMUST00000137312.2
ENSMUST00000106206.8 |
Ndufs5
|
NADH:ubiquinone oxidoreductase core subunit S5 |
| chr15_-_85695291 | 0.87 |
ENSMUST00000125947.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr7_+_45683122 | 0.87 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr15_-_76084035 | 0.85 |
ENSMUST00000054449.14
ENSMUST00000169714.8 ENSMUST00000165453.8 |
Plec
|
plectin |
| chr16_-_20972750 | 0.84 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr9_-_71499594 | 0.83 |
ENSMUST00000166112.2
|
Myzap
|
myocardial zonula adherens protein |
| chr15_-_94302139 | 0.82 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr4_-_116228921 | 0.81 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr4_-_138053603 | 0.80 |
ENSMUST00000030536.13
|
Pink1
|
PTEN induced putative kinase 1 |
| chr15_-_85695810 | 0.79 |
ENSMUST00000144067.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr17_+_48080113 | 0.77 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr5_-_124233812 | 0.77 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr15_-_54953819 | 0.76 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr10_-_5144699 | 0.76 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr7_-_30523191 | 0.76 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr19_-_33739351 | 0.75 |
ENSMUST00000025694.7
|
Lipo3
|
lipase, member O3 |
| chr12_-_115459678 | 0.75 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr15_-_12549350 | 0.75 |
ENSMUST00000190929.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr1_-_163552693 | 0.74 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr4_-_138053545 | 0.73 |
ENSMUST00000105817.4
|
Pink1
|
PTEN induced putative kinase 1 |
| chr1_+_85856204 | 0.73 |
ENSMUST00000027426.11
|
4933407L21Rik
|
RIKEN cDNA 4933407L21 gene |
| chr16_+_93526987 | 0.72 |
ENSMUST00000227156.2
|
Dop1b
|
DOP1 leucine zipper like protein B |
| chr4_-_155013002 | 0.72 |
ENSMUST00000152687.8
ENSMUST00000137803.8 ENSMUST00000145296.2 |
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr19_+_29902506 | 0.71 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr3_-_146518706 | 0.71 |
ENSMUST00000102515.10
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr5_-_94075731 | 0.70 |
ENSMUST00000179824.2
|
Pramel36
|
PRAME like 36 |
| chr12_+_33003882 | 0.65 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chr18_+_37847792 | 0.63 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr4_-_149211145 | 0.62 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chr3_-_126918491 | 0.61 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr5_-_139805661 | 0.60 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr17_+_34524841 | 0.60 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr17_+_34524884 | 0.59 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr2_+_3337194 | 0.58 |
ENSMUST00000115089.2
ENSMUST00000228935.2 |
Acbd7
|
acyl-Coenzyme A binding domain containing 7 |
| chr6_-_121058551 | 0.58 |
ENSMUST00000077159.8
ENSMUST00000207889.2 ENSMUST00000238957.2 ENSMUST00000238920.2 ENSMUST00000238968.2 |
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr10_+_87982854 | 0.57 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr2_-_129139125 | 0.56 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr2_-_174314672 | 0.56 |
ENSMUST00000117442.8
ENSMUST00000141100.2 ENSMUST00000120822.2 |
Prelid3b
|
PRELI domain containing 3B |
| chr2_+_85519775 | 0.56 |
ENSMUST00000054868.2
|
Olfr1008
|
olfactory receptor 1008 |
| chr10_+_80662490 | 0.56 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr17_+_34258411 | 0.55 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr2_-_174314741 | 0.55 |
ENSMUST00000016401.15
|
Prelid3b
|
PRELI domain containing 3B |
| chr7_+_30411634 | 0.55 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr5_-_131567526 | 0.54 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr13_+_108452866 | 0.53 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chr7_+_120682472 | 0.52 |
ENSMUST00000171880.3
ENSMUST00000047025.15 ENSMUST00000170106.2 ENSMUST00000168311.8 |
Otoa
|
otoancorin |
| chr5_-_36739724 | 0.52 |
ENSMUST00000031094.15
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr13_+_108452930 | 0.51 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr3_+_138047536 | 0.51 |
ENSMUST00000199673.6
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr9_-_107956474 | 0.49 |
ENSMUST00000162355.8
ENSMUST00000047746.13 ENSMUST00000174504.8 ENSMUST00000178267.8 ENSMUST00000160649.8 |
Rnf123
|
ring finger protein 123 |
| chr9_-_62417780 | 0.48 |
ENSMUST00000164246.9
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr2_-_120561983 | 0.47 |
ENSMUST00000110701.8
ENSMUST00000110700.2 |
Cdan1
|
congenital dyserythropoietic anemia, type I (human) |
| chr8_-_70892204 | 0.46 |
ENSMUST00000076615.6
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr15_+_34837501 | 0.46 |
ENSMUST00000072868.5
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr1_-_5140504 | 0.46 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr5_+_136145485 | 0.45 |
ENSMUST00000111127.8
ENSMUST00000041366.14 ENSMUST00000111129.2 |
Polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr4_-_82778066 | 0.45 |
ENSMUST00000107239.8
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr6_-_142750192 | 0.45 |
ENSMUST00000111768.3
|
Sult6b2
|
sulfotransferase family 6B, member 2 |
| chrX_+_150127171 | 0.45 |
ENSMUST00000073364.6
|
Fam120c
|
family with sequence similarity 120, member C |
| chr6_-_78355834 | 0.44 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr10_-_82328442 | 0.44 |
ENSMUST00000177934.3
|
Gm1553
|
predicted gene 1553 |
| chr17_-_46513499 | 0.43 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr10_+_80136830 | 0.42 |
ENSMUST00000140828.8
ENSMUST00000138909.8 |
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr3_+_105811712 | 0.42 |
ENSMUST00000000574.3
|
Adora3
|
adenosine A3 receptor |
| chr6_-_70120881 | 0.42 |
ENSMUST00000103380.3
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr7_-_139982831 | 0.42 |
ENSMUST00000080153.4
ENSMUST00000216053.2 ENSMUST00000217167.2 |
Olfr531
|
olfactory receptor 531 |
| chr7_-_25176959 | 0.41 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr10_+_87982916 | 0.41 |
ENSMUST00000169309.3
|
Nup37
|
nucleoporin 37 |
| chr2_-_34716199 | 0.41 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr14_+_32935983 | 0.41 |
ENSMUST00000165792.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr5_-_121147593 | 0.40 |
ENSMUST00000079204.9
|
Rph3a
|
rabphilin 3A |
| chr16_-_58930996 | 0.40 |
ENSMUST00000076262.4
|
Olfr193
|
olfactory receptor 193 |
| chr4_+_152270510 | 0.40 |
ENSMUST00000167926.8
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr5_-_121148143 | 0.39 |
ENSMUST00000202406.4
ENSMUST00000200792.2 |
Rph3a
|
rabphilin 3A |
| chr4_-_114991174 | 0.38 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr14_+_53505374 | 0.38 |
ENSMUST00000199112.2
|
Trav15n-2
|
T cell receptor alpha variable 15N-2 |
| chr3_+_138058139 | 0.37 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr2_+_155078522 | 0.37 |
ENSMUST00000150602.2
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr2_-_111820618 | 0.36 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr19_-_6910922 | 0.36 |
ENSMUST00000235248.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr19_-_11852453 | 0.36 |
ENSMUST00000213954.2
ENSMUST00000217617.2 |
Olfr1419
|
olfactory receptor 1419 |
| chr17_+_25105617 | 0.36 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr4_-_114991478 | 0.35 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr1_-_93029532 | 0.35 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A |
| chr11_+_78717398 | 0.35 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr2_+_155078449 | 0.34 |
ENSMUST00000109682.9
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr7_-_137012444 | 0.34 |
ENSMUST00000120340.2
ENSMUST00000117404.8 ENSMUST00000068996.13 |
9430038I01Rik
|
RIKEN cDNA 9430038I01 gene |
| chr2_+_30282414 | 0.33 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr19_-_5660057 | 0.33 |
ENSMUST00000236229.2
ENSMUST00000235701.2 ENSMUST00000236264.2 |
Kat5
|
K(lysine) acetyltransferase 5 |
| chr4_+_152270636 | 0.33 |
ENSMUST00000030779.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr18_-_62313019 | 0.33 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr13_+_41267783 | 0.32 |
ENSMUST00000124093.3
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr9_+_85724913 | 0.32 |
ENSMUST00000098500.5
|
Tpbg
|
trophoblast glycoprotein |
| chr2_+_130975417 | 0.32 |
ENSMUST00000110225.2
|
Gm11037
|
predicted gene 11037 |
| chr17_-_13070780 | 0.31 |
ENSMUST00000162389.2
ENSMUST00000162119.8 ENSMUST00000159223.8 |
Mas1
|
MAS1 oncogene |
| chr1_-_93029547 | 0.31 |
ENSMUST00000112958.9
ENSMUST00000186861.2 ENSMUST00000171556.8 |
Kif1a
|
kinesin family member 1A |
| chr6_-_83302890 | 0.31 |
ENSMUST00000204472.2
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr7_+_83234118 | 0.31 |
ENSMUST00000039317.14
ENSMUST00000164944.2 |
Tmc3
|
transmembrane channel-like gene family 3 |
| chr2_-_121337120 | 0.31 |
ENSMUST00000089926.6
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr7_+_28140450 | 0.29 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr6_+_41258194 | 0.29 |
ENSMUST00000191646.6
ENSMUST00000103282.3 |
Trbv30
|
T cell receptor beta, variable 30 |
| chr6_+_82379768 | 0.29 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr4_+_132903646 | 0.29 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.4 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 1.9 | 9.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.2 | 4.8 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.2 | 3.5 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 1.0 | 16.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 6.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 3.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 1.5 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 1.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.4 | 1.6 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 3.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 1.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.2 | 1.9 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 2.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 3.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 17.6 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.2 | 2.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 4.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.8 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.2 | 0.8 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 1.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 0.7 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.9 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 | 1.9 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.2 | 1.5 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.2 | 0.5 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 0.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 6.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 | 5.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.8 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.7 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 1.6 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.7 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 2.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.3 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.1 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 2.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.8 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 | 0.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 1.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.9 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.1 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 0.8 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.2 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.0 | 0.7 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 1.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.8 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 6.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.5 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 3.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 2.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.5 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 2.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.4 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 14.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.0 | 9.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.6 | 1.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 2.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 13.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 6.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 6.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 4.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.5 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 7.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.0 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 4.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 6.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.2 | 15.4 | GO:0071253 | connexin binding(GO:0071253) |
| 1.1 | 3.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 1.0 | 3.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.6 | 1.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.5 | 1.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 3.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 2.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 4.8 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 6.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 6.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.2 | 1.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.5 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 0.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 3.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.8 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.5 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 0.4 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 6.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.8 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.1 | GO:0097506 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 2.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 23.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 2.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.2 | 3.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 7.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 6.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 16.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 4.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 2.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |