Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
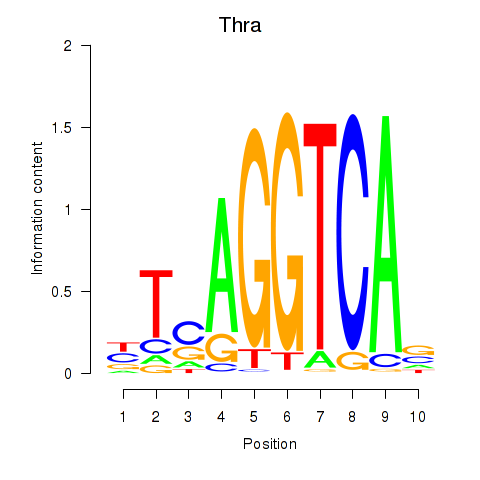
Results for Thra
Z-value: 1.14
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSMUSG00000058756.14 | Thra |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | mm39_v1_chr11_+_98632631_98632642 | -0.47 | 3.0e-05 | Click! |
Activity profile of Thra motif
Sorted Z-values of Thra motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Thra
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_84817547 | 9.61 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr11_+_69856222 | 9.03 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr7_+_141995545 | 7.31 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_+_19144950 | 6.83 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr9_+_122980006 | 6.75 |
ENSMUST00000026890.6
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr7_-_142050663 | 6.71 |
ENSMUST00000238347.2
|
Prr33
|
proline rich 33 |
| chr16_-_10608766 | 5.57 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr7_-_140793990 | 5.17 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chr15_+_102353802 | 4.79 |
ENSMUST00000023809.11
ENSMUST00000229278.2 ENSMUST00000229566.2 |
Amhr2
|
anti-Mullerian hormone type 2 receptor |
| chr7_+_75879603 | 4.78 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr12_+_112645237 | 4.74 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr7_+_45164005 | 4.71 |
ENSMUST00000210532.2
ENSMUST00000085331.14 ENSMUST00000210299.2 |
Tulp2
|
tubby-like protein 2 |
| chr11_-_70146156 | 4.70 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr18_+_11052458 | 4.59 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr15_-_78413780 | 4.45 |
ENSMUST00000229185.2
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr15_-_78413816 | 4.40 |
ENSMUST00000023075.9
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr5_-_21629661 | 4.33 |
ENSMUST00000115245.8
ENSMUST00000030552.7 |
Ccdc146
|
coiled-coil domain containing 146 |
| chr2_+_152753231 | 4.14 |
ENSMUST00000028970.8
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr15_+_102927366 | 3.99 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr1_+_172327569 | 3.87 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr3_+_95071617 | 3.87 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chrX_-_7834057 | 3.85 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chrX_-_142610371 | 3.85 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr1_-_84817022 | 3.84 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr4_+_94627513 | 3.82 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr4_+_126042250 | 3.76 |
ENSMUST00000106150.3
|
Eva1b
|
eva-1 homolog B (C. elegans) |
| chr5_-_116560916 | 3.76 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr9_+_110848339 | 3.74 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr7_-_30755007 | 3.67 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_+_100122192 | 3.65 |
ENSMUST00000032958.14
ENSMUST00000107059.2 |
Ucp3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr1_-_156936197 | 3.64 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr1_+_159871943 | 3.63 |
ENSMUST00000163892.8
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr6_+_90279064 | 3.54 |
ENSMUST00000167550.4
|
BC048671
|
cDNA sequence BC048671 |
| chrX_+_100492684 | 3.49 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr4_-_141327146 | 3.49 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chrX_-_56384089 | 3.33 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_140231618 | 3.31 |
ENSMUST00000122367.8
ENSMUST00000120133.2 |
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr7_-_100164007 | 3.30 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr14_-_70445086 | 3.22 |
ENSMUST00000022682.6
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr7_+_101879176 | 3.16 |
ENSMUST00000120119.9
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr1_-_167294349 | 3.13 |
ENSMUST00000036643.6
|
Lrrc52
|
leucine rich repeat containing 52 |
| chrX_-_165368675 | 3.12 |
ENSMUST00000000412.3
|
Egfl6
|
EGF-like-domain, multiple 6 |
| chr8_-_73197616 | 3.07 |
ENSMUST00000019876.12
|
Calr3
|
calreticulin 3 |
| chr2_-_6217844 | 3.02 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr7_-_4967332 | 3.02 |
ENSMUST00000182214.8
ENSMUST00000032598.14 ENSMUST00000183170.2 |
Sbk2
|
SH3-binding domain kinase family, member 2 |
| chr3_-_95214443 | 2.99 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr7_+_26821266 | 2.95 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr6_+_22288220 | 2.94 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chrX_+_85235370 | 2.89 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr12_-_112893382 | 2.84 |
ENSMUST00000075827.5
|
Jag2
|
jagged 2 |
| chr19_+_42024439 | 2.83 |
ENSMUST00000238137.2
ENSMUST00000026172.3 |
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr2_+_152873772 | 2.79 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr11_+_75541324 | 2.79 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
| chr8_+_72889073 | 2.74 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr19_-_46033353 | 2.73 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr7_+_26819334 | 2.68 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr1_-_84817000 | 2.57 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr7_+_18817767 | 2.56 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr7_-_119694400 | 2.55 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr1_-_190897012 | 2.54 |
ENSMUST00000171798.2
|
Fam71a
|
family with sequence similarity 71, member A |
| chr14_+_79718604 | 2.50 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr4_+_42969934 | 2.49 |
ENSMUST00000190902.2
ENSMUST00000030163.12 |
1700022I11Rik
|
RIKEN cDNA 1700022I11 gene |
| chr8_-_92528543 | 2.48 |
ENSMUST00000093312.6
|
Irx3
|
Iroquois related homeobox 3 |
| chr13_+_51325058 | 2.47 |
ENSMUST00000021828.6
|
Nxnl2
|
nucleoredoxin-like 2 |
| chr6_+_49372463 | 2.40 |
ENSMUST00000024171.14
ENSMUST00000163954.8 ENSMUST00000172459.8 |
Stk31
|
serine threonine kinase 31 |
| chr18_+_82932747 | 2.39 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr17_-_89508103 | 2.39 |
ENSMUST00000035701.6
|
Fshr
|
follicle stimulating hormone receptor |
| chr3_-_108797022 | 2.37 |
ENSMUST00000180063.8
ENSMUST00000053065.8 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr7_-_126574959 | 2.36 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr4_-_41048124 | 2.35 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr19_-_56378459 | 2.32 |
ENSMUST00000040711.15
ENSMUST00000095947.11 ENSMUST00000073536.13 |
Nrap
|
nebulin-related anchoring protein |
| chr16_-_18904240 | 2.32 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr3_+_89970088 | 2.28 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr11_+_83741657 | 2.26 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr19_-_10460238 | 2.25 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr7_-_80052491 | 2.24 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr17_+_35235552 | 2.23 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr8_+_110220614 | 2.23 |
ENSMUST00000034162.8
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr3_+_122523219 | 2.22 |
ENSMUST00000200389.2
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr9_+_107784065 | 2.20 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr4_-_41275091 | 2.20 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr11_+_83741689 | 2.17 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr16_+_24540599 | 2.16 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_122144125 | 2.15 |
ENSMUST00000147788.8
ENSMUST00000154412.2 ENSMUST00000110537.8 ENSMUST00000148417.8 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr3_-_94693780 | 2.14 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr4_-_82939330 | 2.11 |
ENSMUST00000071708.12
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr1_+_134121170 | 2.09 |
ENSMUST00000038445.13
ENSMUST00000191577.2 |
Mybph
|
myosin binding protein H |
| chr11_+_29497950 | 2.08 |
ENSMUST00000020753.4
ENSMUST00000208530.2 |
Clhc1
|
clathrin heavy chain linker domain containing 1 |
| chr4_+_143076327 | 2.07 |
ENSMUST00000052458.3
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr1_+_131678223 | 2.05 |
ENSMUST00000147800.2
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr9_-_107486381 | 2.02 |
ENSMUST00000102531.7
ENSMUST00000102530.8 ENSMUST00000195057.2 ENSMUST00000102532.10 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr10_-_17823736 | 2.01 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr15_-_76193955 | 1.98 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr17_-_71575584 | 1.98 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr1_+_86354045 | 1.98 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr4_-_137493785 | 1.97 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr11_+_85061922 | 1.97 |
ENSMUST00000018623.4
|
1700125H20Rik
|
RIKEN cDNA 1700125H20 gene |
| chr5_-_137623331 | 1.94 |
ENSMUST00000031732.14
|
Fbxo24
|
F-box protein 24 |
| chr5_+_34527230 | 1.90 |
ENSMUST00000180376.8
|
Fam193a
|
family with sequence homology 193, member A |
| chr10_-_62363217 | 1.90 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr7_+_18962301 | 1.86 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr4_-_141327253 | 1.84 |
ENSMUST00000147785.8
|
Fblim1
|
filamin binding LIM protein 1 |
| chr15_-_5273659 | 1.84 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr11_-_40624200 | 1.80 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr12_-_98703664 | 1.79 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr3_+_95067759 | 1.78 |
ENSMUST00000131742.8
ENSMUST00000090823.8 ENSMUST00000090821.10 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr11_+_101875095 | 1.75 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr19_-_56378309 | 1.75 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr4_-_140501507 | 1.74 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr11_+_53991750 | 1.74 |
ENSMUST00000093107.12
ENSMUST00000019050.12 ENSMUST00000174616.8 ENSMUST00000129499.8 ENSMUST00000126840.8 |
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr7_-_4967367 | 1.74 |
ENSMUST00000208109.2
|
Sbk2
|
SH3-binding domain kinase family, member 2 |
| chr18_+_60907668 | 1.72 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr4_+_44300876 | 1.69 |
ENSMUST00000045607.12
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr11_+_68484879 | 1.66 |
ENSMUST00000102612.2
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr19_-_20931566 | 1.65 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr4_-_42665763 | 1.63 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr17_-_35077089 | 1.62 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr18_+_60907698 | 1.61 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr4_+_57821050 | 1.61 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr4_-_42168603 | 1.61 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr17_+_35844091 | 1.60 |
ENSMUST00000025273.9
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr5_-_134975773 | 1.59 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr3_+_32791139 | 1.58 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr6_+_115337899 | 1.58 |
ENSMUST00000171644.8
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr13_-_81781238 | 1.56 |
ENSMUST00000126444.8
ENSMUST00000128585.9 ENSMUST00000146749.2 ENSMUST00000095585.11 |
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr2_+_130248398 | 1.56 |
ENSMUST00000055421.6
|
Tmem239
|
transmembrane 239 |
| chr12_-_57592907 | 1.55 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr10_+_61556371 | 1.54 |
ENSMUST00000080099.6
|
Aifm2
|
apoptosis-inducing factor, mitochondrion-associated 2 |
| chr17_+_85928459 | 1.50 |
ENSMUST00000162695.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr10_-_62363192 | 1.47 |
ENSMUST00000160643.8
|
Srgn
|
serglycin |
| chr10_+_84412490 | 1.47 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr5_-_110987604 | 1.44 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr17_+_80614795 | 1.41 |
ENSMUST00000223878.2
ENSMUST00000068175.6 ENSMUST00000224391.2 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr11_+_100960838 | 1.36 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr10_+_79984097 | 1.36 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr7_-_19530714 | 1.34 |
ENSMUST00000108449.9
ENSMUST00000043822.8 |
Cblc
|
Casitas B-lineage lymphoma c |
| chr15_-_5273645 | 1.33 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr2_-_157121440 | 1.33 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr15_+_73594965 | 1.32 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr4_-_126096551 | 1.31 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr12_-_111679344 | 1.31 |
ENSMUST00000160576.2
|
Bag5
|
BCL2-associated athanogene 5 |
| chr16_+_72460029 | 1.31 |
ENSMUST00000023600.8
|
Robo1
|
roundabout guidance receptor 1 |
| chr2_-_65069383 | 1.31 |
ENSMUST00000155916.8
ENSMUST00000156643.2 |
Cobll1
|
Cobl-like 1 |
| chr3_+_86131970 | 1.29 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr2_+_155223728 | 1.27 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr5_+_110987839 | 1.26 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr7_-_140676623 | 1.26 |
ENSMUST00000209352.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_+_3339059 | 1.25 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr4_+_131600918 | 1.25 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chrX_+_72760183 | 1.24 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr2_-_151318073 | 1.24 |
ENSMUST00000080132.3
|
4921509C19Rik
|
RIKEN cDNA 4921509C19 gene |
| chr1_-_163822336 | 1.24 |
ENSMUST00000097493.10
ENSMUST00000045876.8 |
BC055324
|
cDNA sequence BC055324 |
| chr7_+_3339077 | 1.23 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr9_+_110643054 | 1.23 |
ENSMUST00000098345.3
|
Prss44
|
protease, serine 44 |
| chr7_+_12246415 | 1.23 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
| chr10_-_81436671 | 1.20 |
ENSMUST00000151858.8
ENSMUST00000142948.8 ENSMUST00000072020.9 |
Tle6
|
transducin-like enhancer of split 6 |
| chr1_-_134006847 | 1.20 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr7_+_28140352 | 1.20 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr11_+_53992054 | 1.18 |
ENSMUST00000135653.8
|
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr15_+_73595012 | 1.16 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr14_-_118289557 | 1.16 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr3_+_63202687 | 1.16 |
ENSMUST00000194836.6
ENSMUST00000191633.6 |
Mme
|
membrane metallo endopeptidase |
| chr13_-_65200204 | 1.16 |
ENSMUST00000222769.2
|
Prss47
|
protease, serine 47 |
| chr17_+_79919267 | 1.15 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr7_-_140676596 | 1.15 |
ENSMUST00000209199.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr4_-_126096376 | 1.15 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr9_+_44152029 | 1.14 |
ENSMUST00000217510.2
ENSMUST00000216632.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_-_13061333 | 1.13 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr17_+_23898223 | 1.12 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr13_+_95012107 | 1.10 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr16_+_32249713 | 1.09 |
ENSMUST00000115137.8
ENSMUST00000079791.11 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr11_+_83743746 | 1.09 |
ENSMUST00000108113.3
|
Hnf1b
|
HNF1 homeobox B |
| chr1_-_84817976 | 1.09 |
ENSMUST00000190067.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr4_-_128856213 | 1.08 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr7_+_12656217 | 1.05 |
ENSMUST00000108539.8
ENSMUST00000004554.14 ENSMUST00000147435.8 ENSMUST00000137329.4 |
Rps5
|
ribosomal protein S5 |
| chr5_-_110987441 | 1.03 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr19_-_6835538 | 1.03 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr16_+_23338960 | 1.03 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr2_+_155224105 | 1.03 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr8_+_106434901 | 1.02 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr7_+_100970435 | 1.01 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr4_+_54947976 | 1.01 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr5_+_53747556 | 1.01 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_+_107194446 | 1.01 |
ENSMUST00000040056.15
ENSMUST00000208956.2 |
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_-_111679618 | 1.00 |
ENSMUST00000054636.7
|
Bag5
|
BCL2-associated athanogene 5 |
| chr11_+_51541728 | 1.00 |
ENSMUST00000117859.8
ENSMUST00000064493.6 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr3_+_63202940 | 0.99 |
ENSMUST00000194150.6
|
Mme
|
membrane metallo endopeptidase |
| chr8_+_73197718 | 0.98 |
ENSMUST00000064853.13
ENSMUST00000121902.2 |
1700030K09Rik
|
RIKEN cDNA 1700030K09 gene |
| chr12_+_111679689 | 0.98 |
ENSMUST00000040519.12
ENSMUST00000163220.10 ENSMUST00000162316.2 |
Coa8
|
cytochrome c oxidase assembly factor 8 |
| chr12_-_111679379 | 0.96 |
ENSMUST00000160825.2
ENSMUST00000162953.2 |
Bag5
|
BCL2-associated athanogene 5 |
| chr10_-_87982732 | 0.93 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr10_+_87982854 | 0.92 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr14_+_47069667 | 0.91 |
ENSMUST00000140114.3
ENSMUST00000133989.8 |
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr4_-_58912678 | 0.91 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr7_+_28140450 | 0.91 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr7_+_106413336 | 0.90 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chr18_-_64794338 | 0.89 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr17_-_24863956 | 0.87 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr8_-_106434565 | 0.86 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr9_+_44151962 | 0.86 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 1.4 | 5.5 | GO:0061215 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.3 | 3.9 | GO:0030221 | basophil differentiation(GO:0030221) |
| 1.2 | 7.5 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 1.1 | 6.8 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 1.1 | 3.2 | GO:1990773 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 1.0 | 4.1 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.9 | 4.7 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.9 | 3.7 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.9 | 2.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.8 | 3.4 | GO:0033368 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.8 | 3.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.8 | 2.4 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.8 | 3.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.8 | 6.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.7 | 2.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.6 | 2.9 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 1.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 2.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.5 | 4.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.5 | 2.5 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.5 | 2.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.5 | 2.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.5 | 2.4 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.5 | 3.3 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.4 | 1.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.4 | 9.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.4 | 1.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.4 | 1.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.4 | 1.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 2.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.3 | 3.8 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.7 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 2.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 3.7 | GO:1903797 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.3 | 2.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 2.9 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 1.3 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.3 | 5.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 2.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 2.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 4.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 1.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 1.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 5.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 1.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 7.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 1.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.6 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.2 | 6.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.4 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 0.7 | GO:0009414 | response to water deprivation(GO:0009414) cellular response to water deprivation(GO:0042631) |
| 0.2 | 1.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.5 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.2 | 3.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.5 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 4.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 2.8 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 2.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 5.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 1.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.9 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.5 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 6.8 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 2.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 3.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 4.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.6 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 1.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 2.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.1 | 1.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 1.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.2 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 1.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 2.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.8 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 1.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 2.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.0 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.9 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 2.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 4.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.7 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 2.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.0 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 2.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 2.1 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 2.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 3.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.5 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 4.8 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 2.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 1.0 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.5 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 6.8 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.2 | GO:0008306 | associative learning(GO:0008306) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.8 | GO:0001652 | granular component(GO:0001652) |
| 0.6 | 3.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.6 | 2.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.6 | 2.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 4.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 7.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 11.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 1.6 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.3 | 3.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 2.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 2.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.5 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.2 | 2.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.7 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 5.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 0.6 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 1.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 10.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 11.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 5.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.1 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 3.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 2.8 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 1.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 17.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 3.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 4.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 7.0 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.8 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.2 | 4.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 1.0 | 6.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 7.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 3.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 4.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 2.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.5 | 2.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.5 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 3.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 2.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 1.5 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 2.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 3.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 5.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 0.8 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.2 | 0.7 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 2.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 5.6 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 7.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 3.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 9.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 5.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.8 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 3.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 1.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.0 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 5.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 2.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 4.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 4.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 4.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 3.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 3.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 5.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 2.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 3.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 5.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 8.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 25.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 9.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 10.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 7.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 6.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 2.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 8.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 11.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 10.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 6.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 9.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 3.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 7.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |