Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
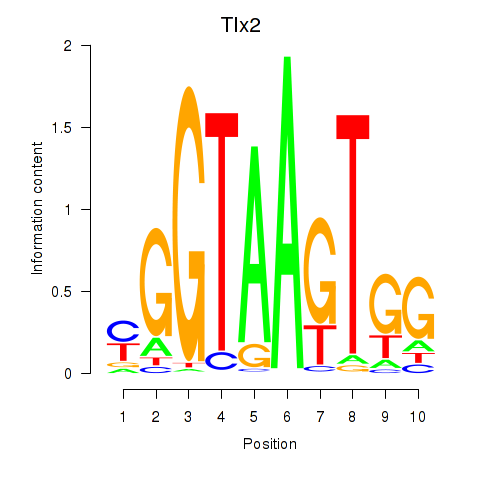
Results for Tlx2
Z-value: 1.67
Transcription factors associated with Tlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx2
|
ENSMUSG00000068327.6 | Tlx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tlx2 | mm39_v1_chr6_-_83047206_83047361 | 0.02 | 8.6e-01 | Click! |
Activity profile of Tlx2 motif
Sorted Z-values of Tlx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11243530 | 15.02 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr3_+_90576285 | 10.37 |
ENSMUST00000069927.10
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr17_+_35413415 | 10.33 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr3_-_90603013 | 9.97 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr6_+_41520150 | 9.60 |
ENSMUST00000103295.2
|
Trbj2-3
|
T cell receptor beta joining 2-3 |
| chr5_-_105198913 | 9.57 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr11_-_52173391 | 8.48 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr7_-_44181477 | 8.22 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chrX_-_100310959 | 8.05 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr19_-_46033353 | 7.97 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr6_+_41520379 | 7.84 |
ENSMUST00000103297.2
|
Trbj2-5
|
T cell receptor beta joining 2-5 |
| chr16_-_36486429 | 7.69 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr6_+_41519884 | 7.56 |
ENSMUST00000103294.2
|
Trbj2-2
|
T cell receptor beta joining 2-2 |
| chr16_-_19019100 | 7.42 |
ENSMUST00000103749.3
|
Iglc2
|
immunoglobulin lambda constant 2 |
| chr6_+_41520287 | 7.33 |
ENSMUST00000103296.2
|
Trbj2-4
|
T cell receptor beta joining 2-4 |
| chr7_-_126736979 | 7.30 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr10_+_79852487 | 7.22 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr10_+_79852750 | 6.99 |
ENSMUST00000105373.8
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr12_+_24758968 | 6.89 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr19_-_9065309 | 6.77 |
ENSMUST00000025554.3
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr1_+_61017057 | 6.69 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr4_+_138503046 | 6.68 |
ENSMUST00000030528.9
|
Pla2g2d
|
phospholipase A2, group IID |
| chr7_+_43057611 | 6.63 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr12_-_113324852 | 6.29 |
ENSMUST00000223179.2
ENSMUST00000103423.3 |
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr19_-_4241034 | 6.24 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr16_+_48692976 | 6.21 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr11_+_106642052 | 6.18 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr19_-_4240984 | 6.13 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr11_+_95728042 | 6.13 |
ENSMUST00000107712.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr5_-_137856280 | 6.08 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr4_-_129452148 | 6.04 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr2_-_126341757 | 6.02 |
ENSMUST00000040128.12
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr4_-_129452180 | 5.99 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr6_+_117145496 | 5.90 |
ENSMUST00000112866.8
ENSMUST00000112871.8 ENSMUST00000073043.5 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr12_-_113552322 | 5.85 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr15_-_97629209 | 5.59 |
ENSMUST00000100249.10
|
Endou
|
endonuclease, polyU-specific |
| chr3_+_87283687 | 5.56 |
ENSMUST00000163661.8
ENSMUST00000072480.9 |
Fcrl1
|
Fc receptor-like 1 |
| chr7_+_28136861 | 5.55 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr16_+_49675682 | 5.50 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr1_+_172327569 | 5.45 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr8_+_13087805 | 5.40 |
ENSMUST00000128418.8
ENSMUST00000152034.2 |
F10
|
coagulation factor X |
| chr8_+_81220410 | 5.38 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr1_-_45964730 | 5.30 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr9_+_7764042 | 5.29 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr16_+_49675969 | 5.29 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr19_+_6451667 | 5.28 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr11_+_106642079 | 5.21 |
ENSMUST00000183111.8
ENSMUST00000106794.9 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr16_+_49676130 | 5.18 |
ENSMUST00000230641.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chrX_+_73314418 | 5.17 |
ENSMUST00000008826.14
ENSMUST00000151702.8 ENSMUST00000074085.12 ENSMUST00000135690.2 |
Rpl10
|
ribosomal protein L10 |
| chr5_-_137834470 | 5.12 |
ENSMUST00000110980.2
ENSMUST00000058897.11 ENSMUST00000199028.2 |
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr12_+_24758240 | 5.10 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_-_60055082 | 5.01 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr12_+_24758724 | 4.97 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr1_+_172327812 | 4.97 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr10_+_79824418 | 4.95 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr2_-_118377500 | 4.91 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr10_+_43455157 | 4.88 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr8_-_25592385 | 4.78 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr3_-_9898713 | 4.76 |
ENSMUST00000161949.8
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr8_+_84682136 | 4.67 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr17_+_8454939 | 4.65 |
ENSMUST00000164411.10
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr17_+_18108102 | 4.65 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr7_+_101034604 | 4.63 |
ENSMUST00000130016.8
ENSMUST00000134143.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr3_+_87283767 | 4.60 |
ENSMUST00000194786.6
ENSMUST00000191666.2 |
Fcrl1
|
Fc receptor-like 1 |
| chr12_+_98234884 | 4.60 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr17_+_18108086 | 4.56 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr17_-_74257164 | 4.51 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr15_-_100585789 | 4.47 |
ENSMUST00000023775.9
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr2_+_164897547 | 4.46 |
ENSMUST00000017799.12
ENSMUST00000073707.9 |
Cd40
|
CD40 antigen |
| chr3_+_87283748 | 4.40 |
ENSMUST00000167200.7
|
Fcrl1
|
Fc receptor-like 1 |
| chr2_+_164897583 | 4.35 |
ENSMUST00000081310.11
ENSMUST00000140951.8 |
Cd40
|
CD40 antigen |
| chr15_+_75468473 | 4.35 |
ENSMUST00000189944.7
ENSMUST00000023243.11 |
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr8_+_66070661 | 4.32 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr11_+_115353290 | 4.27 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr19_-_4213347 | 4.25 |
ENSMUST00000025749.15
|
Rps6kb2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr8_-_25591737 | 4.22 |
ENSMUST00000098866.11
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr1_+_34511793 | 4.22 |
ENSMUST00000188972.3
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr18_+_60509101 | 4.13 |
ENSMUST00000032473.7
ENSMUST00000066912.13 |
Iigp1
|
interferon inducible GTPase 1 |
| chr11_+_115790768 | 4.09 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr16_-_19525122 | 4.06 |
ENSMUST00000081880.7
|
Lamp3
|
lysosomal-associated membrane protein 3 |
| chr1_-_80642969 | 4.05 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr2_-_117173312 | 4.02 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr5_-_105491795 | 4.01 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chr13_-_49473695 | 4.00 |
ENSMUST00000110086.2
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr16_+_44586085 | 3.97 |
ENSMUST00000057488.15
|
Cd200r1
|
CD200 receptor 1 |
| chr10_-_62343516 | 3.96 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr1_-_36312482 | 3.93 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr14_+_53791444 | 3.91 |
ENSMUST00000198297.2
|
Trav14-1
|
T cell receptor alpha variable 14-1 |
| chr1_+_34498836 | 3.91 |
ENSMUST00000027302.14
ENSMUST00000190122.2 |
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr2_-_117173428 | 3.85 |
ENSMUST00000102534.11
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr8_-_106660470 | 3.83 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr4_-_63779562 | 3.81 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr2_+_127967951 | 3.79 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chrX_+_99669343 | 3.78 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr10_+_79722081 | 3.75 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr10_+_98943999 | 3.73 |
ENSMUST00000161240.4
|
Galnt4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr7_-_46365108 | 3.69 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr2_+_84818538 | 3.67 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr3_+_103767581 | 3.64 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr14_+_53941464 | 3.61 |
ENSMUST00000103664.6
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr2_-_117173190 | 3.59 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr14_+_54444554 | 3.58 |
ENSMUST00000103730.2
|
Traj11
|
T cell receptor alpha joining 11 |
| chr14_+_53859114 | 3.57 |
ENSMUST00000103657.6
|
Trav12-3
|
T cell receptor alpha variable 12-3 |
| chr17_-_31363245 | 3.55 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr17_+_8454862 | 3.55 |
ENSMUST00000231340.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr6_-_99497900 | 3.54 |
ENSMUST00000176565.8
|
Foxp1
|
forkhead box P1 |
| chr3_-_9898676 | 3.53 |
ENSMUST00000108384.9
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr19_-_7218363 | 3.50 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chrX_+_158491589 | 3.47 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr14_+_53059379 | 3.43 |
ENSMUST00000197754.2
|
Trav6d-6
|
T cell receptor alpha variable 6D-6 |
| chr3_-_59038634 | 3.42 |
ENSMUST00000200358.2
ENSMUST00000197220.2 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr17_+_34816826 | 3.41 |
ENSMUST00000015596.10
ENSMUST00000174496.9 ENSMUST00000173992.8 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr7_+_82297803 | 3.40 |
ENSMUST00000141726.8
ENSMUST00000179489.8 ENSMUST00000039881.4 |
Efl1
|
elongation factor like GTPase 1 |
| chr13_-_19521337 | 3.39 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr9_+_107957621 | 3.36 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr6_-_70149254 | 3.33 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr11_+_115790951 | 3.29 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr14_+_53698556 | 3.29 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr14_+_54394723 | 3.28 |
ENSMUST00000103687.2
|
Traj58
|
T cell receptor alpha joining 58 |
| chr9_+_123902143 | 3.25 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr14_-_55950939 | 3.21 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr13_+_19524136 | 3.21 |
ENSMUST00000103564.3
|
Trgv1
|
T cell receptor gamma, variable 1 |
| chr12_-_32000534 | 3.19 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr14_-_54386718 | 3.18 |
ENSMUST00000103685.3
|
Trdv5
|
T cell receptor delta variable 5 |
| chr5_+_108048367 | 3.14 |
ENSMUST00000082223.13
|
Rpl5
|
ribosomal protein L5 |
| chr17_+_87590308 | 3.13 |
ENSMUST00000041110.12
ENSMUST00000125875.8 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr12_-_113843161 | 3.13 |
ENSMUST00000103451.5
|
Ighv2-9
|
immunoglobulin heavy variable 2-9 |
| chr1_-_80643024 | 3.12 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr6_-_41681273 | 3.11 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr6_+_67838100 | 3.10 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr15_-_74599860 | 3.10 |
ENSMUST00000023261.4
ENSMUST00000190433.2 |
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr6_-_99243455 | 3.09 |
ENSMUST00000113326.9
|
Foxp1
|
forkhead box P1 |
| chr12_+_102249294 | 3.08 |
ENSMUST00000056950.14
|
Rin3
|
Ras and Rab interactor 3 |
| chr5_+_108048697 | 3.08 |
ENSMUST00000153590.2
|
Rpl5
|
ribosomal protein L5 |
| chr5_-_137834444 | 3.07 |
ENSMUST00000197586.2
|
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr19_-_11301919 | 3.06 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr17_-_52140305 | 3.04 |
ENSMUST00000133574.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr9_-_108888284 | 3.03 |
ENSMUST00000112053.2
|
Trex1
|
three prime repair exonuclease 1 |
| chr3_+_68912043 | 3.02 |
ENSMUST00000042901.15
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr6_+_129022843 | 3.00 |
ENSMUST00000032257.10
ENSMUST00000204320.2 |
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F |
| chr11_-_106051533 | 2.96 |
ENSMUST00000106875.2
|
Limd2
|
LIM domain containing 2 |
| chr2_-_129139125 | 2.93 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chrX_-_36253309 | 2.91 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr3_-_20089433 | 2.88 |
ENSMUST00000108321.2
ENSMUST00000012580.13 |
Hps3
|
HPS3, biogenesis of lysosomal organelles complex 2 subunit 1 |
| chr3_-_36529753 | 2.86 |
ENSMUST00000199478.2
|
Anxa5
|
annexin A5 |
| chr15_-_34443054 | 2.86 |
ENSMUST00000142643.2
|
Rpl30
|
ribosomal protein L30 |
| chr3_-_96201248 | 2.86 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr11_-_78875689 | 2.86 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr13_+_102830029 | 2.85 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr14_+_32507920 | 2.85 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr2_+_180098026 | 2.84 |
ENSMUST00000038259.13
|
Slco4a1
|
solute carrier organic anion transporter family, member 4a1 |
| chr9_+_107957640 | 2.82 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr16_+_91184661 | 2.81 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr6_+_38895902 | 2.80 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr14_+_54444010 | 2.80 |
ENSMUST00000103729.2
|
Traj12
|
T cell receptor alpha joining 12 |
| chr19_-_4251589 | 2.78 |
ENSMUST00000237923.2
ENSMUST00000025740.8 ENSMUST00000237723.2 |
Rad9a
|
RAD9 checkpoint clamp component A |
| chr16_-_3535940 | 2.76 |
ENSMUST00000229725.2
ENSMUST00000100222.4 |
Mefv
|
Mediterranean fever |
| chr10_+_79715448 | 2.76 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3 |
| chr7_+_30122685 | 2.75 |
ENSMUST00000046177.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr15_+_103148824 | 2.72 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr6_+_38918327 | 2.70 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr14_+_45225641 | 2.69 |
ENSMUST00000046891.6
|
Ptger2
|
prostaglandin E receptor 2 (subtype EP2) |
| chr4_+_137944546 | 2.69 |
ENSMUST00000130071.2
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr5_+_45827249 | 2.66 |
ENSMUST00000117396.3
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr8_+_67947494 | 2.64 |
ENSMUST00000093470.7
ENSMUST00000163856.3 |
Nat2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr6_+_78382131 | 2.64 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr11_+_101333238 | 2.61 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr14_+_54701594 | 2.59 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr11_-_75918551 | 2.55 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B |
| chr19_+_6108240 | 2.55 |
ENSMUST00000237840.2
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr16_-_3535958 | 2.54 |
ENSMUST00000023180.15
|
Mefv
|
Mediterranean fever |
| chr3_-_92577621 | 2.53 |
ENSMUST00000029530.6
|
Lce1a2
|
late cornified envelope 1A2 |
| chr4_+_130253925 | 2.53 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr11_-_78875657 | 2.53 |
ENSMUST00000073001.5
|
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr11_+_101333115 | 2.51 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr3_-_59038160 | 2.51 |
ENSMUST00000197841.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr16_+_87495792 | 2.51 |
ENSMUST00000026703.6
|
Bach1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr6_-_72367642 | 2.51 |
ENSMUST00000059983.10
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr11_+_44290884 | 2.50 |
ENSMUST00000170513.3
ENSMUST00000102796.10 |
Il12b
|
interleukin 12b |
| chr8_-_106863521 | 2.48 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr7_+_78563964 | 2.48 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr13_-_20008397 | 2.43 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr11_-_5848771 | 2.42 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr1_-_162726234 | 2.40 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr11_+_68936457 | 2.39 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr19_-_7218512 | 2.39 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr1_-_91340884 | 2.37 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr4_+_63274388 | 2.35 |
ENSMUST00000006687.5
|
Orm3
|
orosomucoid 3 |
| chr3_+_92874366 | 2.35 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr9_+_94551929 | 2.35 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr12_-_32000169 | 2.34 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr10_-_34083570 | 2.33 |
ENSMUST00000215547.2
ENSMUST00000048010.9 |
Dse
|
dermatan sulfate epimerase |
| chr14_+_53954133 | 2.32 |
ENSMUST00000103641.6
|
Trav7-6
|
T cell receptor alpha variable 7-6 |
| chr11_-_99501015 | 2.32 |
ENSMUST00000076478.2
|
Gm11937
|
predicted gene 11937 |
| chr7_-_112987770 | 2.31 |
ENSMUST00000079793.7
|
Pth
|
parathyroid hormone |
| chr4_+_137944438 | 2.30 |
ENSMUST00000137851.8
ENSMUST00000165861.8 |
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr1_+_172302925 | 2.29 |
ENSMUST00000027830.5
|
Slamf9
|
SLAM family member 9 |
| chr6_+_68657317 | 2.28 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr8_-_106863423 | 2.28 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chrX_-_50294867 | 2.27 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.3 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 2.7 | 8.2 | GO:1904155 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 2.7 | 8.0 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 2.4 | 7.3 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 2.1 | 6.2 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 2.1 | 6.2 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 1.9 | 5.6 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.8 | 3.6 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.8 | 5.3 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 1.7 | 6.8 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 1.6 | 4.9 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 1.6 | 4.9 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.6 | 4.8 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 1.6 | 14.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 1.6 | 6.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 1.5 | 4.6 | GO:2000451 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 1.5 | 5.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.5 | 16.0 | GO:0008228 | opsonization(GO:0008228) |
| 1.3 | 8.1 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 1.3 | 9.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.3 | 10.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.2 | 3.7 | GO:0070947 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 1.2 | 5.0 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 1.2 | 4.9 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.1 | 3.4 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 1.1 | 10.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 1.1 | 5.4 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 1.1 | 8.5 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 1.1 | 5.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 1.0 | 2.0 | GO:0002649 | regulation of tolerance induction to self antigen(GO:0002649) |
| 1.0 | 4.0 | GO:0033379 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 1.0 | 2.9 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 1.0 | 6.7 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 1.0 | 2.9 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.9 | 12.0 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.9 | 2.8 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.9 | 3.7 | GO:0045575 | basophil activation(GO:0045575) |
| 0.9 | 4.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.9 | 7.9 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.9 | 13.9 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.8 | 2.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.8 | 4.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 2.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.8 | 3.1 | GO:2000878 | dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 0.8 | 14.4 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.7 | 17.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.7 | 2.9 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.7 | 2.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.7 | 2.8 | GO:0060168 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.7 | 8.8 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.7 | 5.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.6 | 4.5 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.6 | 2.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.6 | 11.4 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.6 | 2.5 | GO:0016240 | autophagosome docking(GO:0016240) pancreatic amylase secretion(GO:0036395) zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.6 | 3.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.6 | 1.8 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.6 | 3.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.6 | 1.8 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.6 | 1.8 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.6 | 2.3 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.6 | 3.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 1.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 5.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 1.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 2.1 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.5 | 8.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.5 | 1.6 | GO:0090172 | attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation(GO:0051455) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.5 | 2.6 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.5 | 1.5 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.5 | 4.6 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.5 | 1.5 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.5 | 2.5 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.5 | 3.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.5 | 2.8 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.5 | 0.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.4 | 1.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.4 | 1.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.4 | 4.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.4 | 2.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.4 | 1.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.4 | 6.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.4 | 3.8 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.4 | 0.7 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.4 | 7.8 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.4 | 2.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 1.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.3 | 1.4 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 8.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 1.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.3 | 1.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 3.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 1.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.3 | 3.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 3.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.3 | 0.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 3.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.3 | 1.4 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.3 | 2.8 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.3 | 1.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 0.8 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 1.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 1.3 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 2.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 0.7 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.2 | 2.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 2.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 3.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 1.7 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.2 | 6.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 3.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.2 | 0.7 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.2 | 0.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 6.8 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 1.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 0.7 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 6.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 | 2.8 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 2.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 5.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 3.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.0 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.2 | 5.5 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.2 | 30.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 4.3 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.2 | 8.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 2.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 0.8 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 1.5 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.2 | 1.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.6 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.6 | GO:0036088 | proline catabolic process(GO:0006562) D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 6.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 5.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 1.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 3.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.7 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 1.9 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.2 | 3.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 1.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 1.5 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 2.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 1.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 10.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 1.9 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 5.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 0.6 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.8 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 1.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 7.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 2.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 2.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 3.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.4 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.8 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 6.5 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 4.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 2.7 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 4.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 1.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 1.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.5 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 4.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 2.6 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 1.3 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 3.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 2.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 22.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 2.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.7 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 1.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 3.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 1.5 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.2 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.1 | 1.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 1.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 2.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.7 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 6.4 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 4.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 11.8 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.5 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 0.6 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 3.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 2.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 2.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 2.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.5 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 1.1 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 6.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.8 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 3.3 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.4 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 6.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 6.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.9 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 6.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 1.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.9 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 1.3 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 | 0.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.5 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 1.2 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 1.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.8 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 3.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.7 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.3 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 1.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.3 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.1 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 1.1 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.0 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.7 | GO:0045727 | positive regulation of translation(GO:0045727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 17.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.4 | 15.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.4 | 4.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.0 | 3.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.7 | 8.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.6 | 2.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.5 | 8.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.5 | 5.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.5 | 8.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.4 | 2.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 6.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 6.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.4 | 1.9 | GO:0043293 | apoptosome(GO:0043293) |
| 0.4 | 1.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 10.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 4.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 3.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 23.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 7.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 2.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 0.7 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 9.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 28.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 7.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 2.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 12.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 4.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 5.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 1.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 1.6 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.2 | 2.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 2.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 75.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 3.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 2.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 4.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 5.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 3.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.0 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 1.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 6.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.7 | GO:0042825 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.1 | 4.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 4.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.2 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.1 | 1.0 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 2.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 4.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 3.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 5.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 12.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 3.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 4.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 4.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 7.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 18.5 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 3.0 | 24.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 2.8 | 17.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.4 | 12.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.6 | 8.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 1.5 | 4.6 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 1.5 | 16.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.3 | 5.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 1.2 | 6.2 | GO:0035478 | chylomicron binding(GO:0035478) |
| 1.2 | 5.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.1 | 3.4 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.0 | 8.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.0 | 5.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 1.0 | 11.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.9 | 2.8 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.9 | 4.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.9 | 4.5 | GO:0043546 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.9 | 9.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.8 | 6.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.7 | 6.6 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.7 | 16.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.7 | 9.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 2.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.7 | 2.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.6 | 2.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.6 | 9.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.6 | 5.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 5.9 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.6 | 15.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.6 | 6.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.6 | 2.8 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.5 | 8.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 1.5 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.5 | 10.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.5 | 5.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 1.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.5 | 5.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 3.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.4 | 1.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 2.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 2.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 1.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 2.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 4.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.3 | 1.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.3 | 1.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 1.6 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.3 | 4.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 3.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 1.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 2.0 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 9.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 2.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.3 | 1.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 1.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 13.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 2.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 1.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 11.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.7 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 0.7 | GO:0047312 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.2 | 1.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 7.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 6.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 4.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 28.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 4.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 2.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 7.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 4.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 2.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 2.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 0.9 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.2 | 0.7 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 2.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 2.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 2.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 1.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.6 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 1.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 2.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 3.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 2.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 3.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.9 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 3.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 4.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.8 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.8 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 12.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 8.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 3.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 5.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 5.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 5.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 4.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 4.4 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.1 | 3.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 11.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 9.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 11.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 4.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 8.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 9.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 12.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 5.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.0 | 0.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 19.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 44.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.3 | 5.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 20.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 5.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 5.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 8.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 10.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 7.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 24.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 1.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 8.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 1.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 8.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 1.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 9.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 5.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 6.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 8.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 9.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 3.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 7.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 5.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 9.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 1.4 | 20.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.7 | 17.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.5 | 15.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.5 | 1.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 7.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.5 | 8.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 18.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 5.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 6.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 8.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 8.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 5.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 3.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 2.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 32.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 4.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 6.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 12.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 2.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 12.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 5.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 5.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.2 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 2.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 6.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 3.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 6.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 5.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 4.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 19.3 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.1 | 4.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 8.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 2.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 3.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 10.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 4.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.7 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 6.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.8 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |