Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Trp53
Z-value: 1.69
Transcription factors associated with Trp53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp53
|
ENSMUSG00000059552.14 | Trp53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp53 | mm39_v1_chr11_+_69471219_69471248 | 0.36 | 1.9e-03 | Click! |
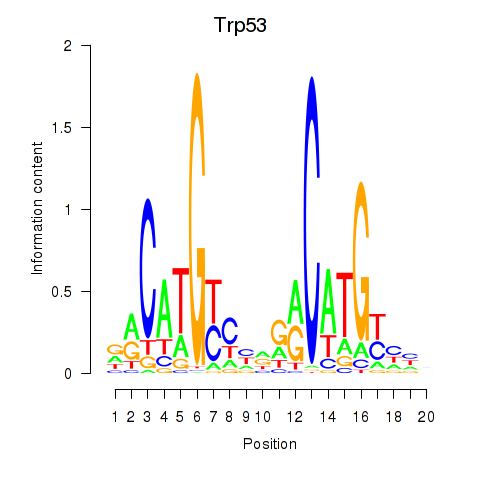
Activity profile of Trp53 motif
Sorted Z-values of Trp53 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp53
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_28426752 | 11.86 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr17_+_35284315 | 11.75 |
ENSMUST00000173207.8
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr5_+_36361360 | 11.66 |
ENSMUST00000052224.6
|
Psapl1
|
prosaposin-like 1 |
| chr2_-_85349362 | 11.59 |
ENSMUST00000099923.2
|
Fads2b
|
fatty acid desaturase 2B |
| chr2_-_69036489 | 11.15 |
ENSMUST00000127243.8
ENSMUST00000149643.2 ENSMUST00000167875.9 ENSMUST00000005365.15 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr1_+_106788873 | 10.31 |
ENSMUST00000086701.13
ENSMUST00000188745.3 ENSMUST00000112730.8 |
Serpinb5
|
serine (or cysteine) peptidase inhibitor, clade B, member 5 |
| chr3_-_132655804 | 10.11 |
ENSMUST00000117164.8
ENSMUST00000093971.5 ENSMUST00000042729.16 |
Npnt
|
nephronectin |
| chr14_-_70412804 | 9.89 |
ENSMUST00000143393.2
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr6_+_41098273 | 9.73 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr3_+_90576285 | 9.66 |
ENSMUST00000069927.10
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr17_+_28426831 | 9.56 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr8_-_106863521 | 9.06 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr2_-_69036472 | 8.93 |
ENSMUST00000112320.8
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chrX_-_72478938 | 8.37 |
ENSMUST00000033738.8
|
Trex2
|
three prime repair exonuclease 2 |
| chr10_-_78554104 | 8.26 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14 |
| chr9_-_76474374 | 8.02 |
ENSMUST00000183437.8
|
Fam83b
|
family with sequence similarity 83, member B |
| chr3_-_144804784 | 8.01 |
ENSMUST00000040465.11
ENSMUST00000198993.2 |
Clca2
|
chloride channel accessory 2 |
| chr8_-_22675773 | 7.78 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr17_-_34109513 | 7.76 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr19_+_41017714 | 7.73 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr11_+_81948649 | 7.70 |
ENSMUST00000000342.3
|
Ccl11
|
chemokine (C-C motif) ligand 11 |
| chr5_-_30278552 | 7.36 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr14_-_56181993 | 7.02 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr7_-_24245419 | 6.88 |
ENSMUST00000011776.8
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr6_-_136918495 | 6.62 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_-_124698805 | 6.52 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_-_136918885 | 6.51 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr19_-_57185808 | 6.47 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_-_111755892 | 6.43 |
ENSMUST00000102720.8
|
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr4_-_135300934 | 6.27 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr14_+_73475335 | 6.25 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr2_-_122144125 | 6.15 |
ENSMUST00000147788.8
ENSMUST00000154412.2 ENSMUST00000110537.8 ENSMUST00000148417.8 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr15_+_79784365 | 6.14 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr1_+_172327569 | 5.94 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr2_-_132787790 | 5.93 |
ENSMUST00000038280.5
|
Fermt1
|
fermitin family member 1 |
| chr16_+_38279289 | 5.93 |
ENSMUST00000099816.3
ENSMUST00000232409.2 |
Cd80
|
CD80 antigen |
| chr9_-_100368841 | 5.88 |
ENSMUST00000098458.4
|
Il20rb
|
interleukin 20 receptor beta |
| chr11_-_3477916 | 5.61 |
ENSMUST00000020718.10
|
Smtn
|
smoothelin |
| chr6_-_136918844 | 5.60 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr7_+_140659038 | 5.56 |
ENSMUST00000159375.8
|
Pkp3
|
plakophilin 3 |
| chr17_+_47747657 | 5.46 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr14_+_55998847 | 5.40 |
ENSMUST00000044554.5
|
Ltb4r2
|
leukotriene B4 receptor 2 |
| chr2_-_163239865 | 5.36 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr3_+_59832635 | 5.31 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr5_+_144127102 | 5.27 |
ENSMUST00000060747.8
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr7_-_24459736 | 5.25 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr6_-_136918671 | 5.18 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr3_-_92481033 | 5.00 |
ENSMUST00000053107.6
|
Ivl
|
involucrin |
| chr14_-_59602882 | 4.90 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr17_+_47747540 | 4.85 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr15_-_102154874 | 4.83 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chr13_-_74956640 | 4.79 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr2_-_17735847 | 4.77 |
ENSMUST00000028080.12
|
Nebl
|
nebulette |
| chr6_-_128868068 | 4.61 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr13_+_24560052 | 4.60 |
ENSMUST00000110391.4
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr17_+_87590308 | 4.60 |
ENSMUST00000041110.12
ENSMUST00000125875.8 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr8_-_89362745 | 4.52 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr9_+_66853343 | 4.51 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr3_+_59914164 | 4.48 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr17_-_35285146 | 4.48 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr12_-_78953703 | 4.40 |
ENSMUST00000021544.8
|
Plek2
|
pleckstrin 2 |
| chr7_+_75105282 | 4.40 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr18_+_66591604 | 4.39 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr3_-_92528480 | 4.39 |
ENSMUST00000170676.3
|
Lce6a
|
late cornified envelope 6A |
| chr11_-_102815910 | 4.28 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr18_-_35781422 | 4.24 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr8_-_94006345 | 4.19 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr2_+_72115981 | 4.17 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr7_+_24584197 | 4.11 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr4_+_114914880 | 4.09 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr4_+_111830119 | 4.09 |
ENSMUST00000106568.8
ENSMUST00000055014.11 ENSMUST00000163281.2 |
Skint7
|
selection and upkeep of intraepithelial T cells 7 |
| chr1_+_133278248 | 4.08 |
ENSMUST00000094556.3
|
Ren1
|
renin 1 structural |
| chr4_-_113144152 | 4.07 |
ENSMUST00000138966.9
|
Skint6
|
selection and upkeep of intraepithelial T cells 6 |
| chr6_+_67586695 | 4.03 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr4_+_155874896 | 4.03 |
ENSMUST00000165000.8
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr13_+_94954202 | 3.98 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr6_+_128991064 | 3.93 |
ENSMUST00000204981.2
|
Clec2f
|
C-type lectin domain family 2, member f |
| chr1_+_88030951 | 3.85 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr1_-_173810310 | 3.80 |
ENSMUST00000000266.9
|
Ifi202b
|
interferon activated gene 202B |
| chr3_+_116653113 | 3.80 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr5_-_139805661 | 3.67 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr10_+_78410803 | 3.66 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr1_-_73967664 | 3.65 |
ENSMUST00000187691.7
|
Tns1
|
tensin 1 |
| chr3_-_75864195 | 3.59 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr1_+_171386752 | 3.59 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr15_+_85744147 | 3.52 |
ENSMUST00000231074.2
|
Gtse1
|
G two S phase expressed protein 1 |
| chr7_+_24584076 | 3.45 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr10_+_110581293 | 3.44 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr1_+_40364752 | 3.42 |
ENSMUST00000193388.2
|
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr6_+_68414401 | 3.38 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr4_-_155141241 | 3.34 |
ENSMUST00000131173.3
|
Plch2
|
phospholipase C, eta 2 |
| chr9_+_61280501 | 3.34 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr15_+_85743887 | 3.25 |
ENSMUST00000170629.3
|
Gtse1
|
G two S phase expressed protein 1 |
| chr1_+_60948149 | 3.23 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr19_-_5399368 | 3.08 |
ENSMUST00000238111.2
|
Cst6
|
cystatin E/M |
| chr7_-_28661751 | 3.03 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr13_-_74956924 | 3.00 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr6_-_116437985 | 2.94 |
ENSMUST00000164547.8
ENSMUST00000170186.2 |
Alox5
|
arachidonate 5-lipoxygenase |
| chr15_+_79784543 | 2.93 |
ENSMUST00000230741.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr9_+_45313913 | 2.92 |
ENSMUST00000214257.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr12_-_70278188 | 2.82 |
ENSMUST00000161083.2
|
Pygl
|
liver glycogen phosphorylase |
| chr7_+_24583994 | 2.82 |
ENSMUST00000108428.8
|
Rps19
|
ribosomal protein S19 |
| chr14_-_59602859 | 2.80 |
ENSMUST00000161031.2
|
Phf11d
|
PHD finger protein 11D |
| chr17_+_36132567 | 2.80 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr11_-_115968373 | 2.80 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr6_-_116438116 | 2.77 |
ENSMUST00000026795.13
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr7_-_141023199 | 2.77 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr17_+_27136065 | 2.76 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr9_-_114811807 | 2.75 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr10_+_75399920 | 2.74 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_+_51328265 | 2.72 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr14_+_74973081 | 2.68 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr4_+_114914607 | 2.52 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr12_-_55539372 | 2.45 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr13_-_98951890 | 2.43 |
ENSMUST00000040340.16
ENSMUST00000179563.8 ENSMUST00000109403.2 |
Fcho2
|
FCH domain only 2 |
| chr1_-_131204422 | 2.39 |
ENSMUST00000159195.2
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr17_-_35077089 | 2.38 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr11_+_73831074 | 2.38 |
ENSMUST00000092917.4
|
Olfr23
|
olfactory receptor 23 |
| chrX_+_73298388 | 2.38 |
ENSMUST00000119197.8
ENSMUST00000088313.5 |
Emd
|
emerin |
| chr5_-_138270995 | 2.31 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr9_+_121946321 | 2.30 |
ENSMUST00000119215.9
ENSMUST00000146832.8 ENSMUST00000118886.9 ENSMUST00000120173.9 ENSMUST00000139181.2 |
Snrk
|
SNF related kinase |
| chr14_+_53797089 | 2.30 |
ENSMUST00000200101.2
ENSMUST00000103653.3 |
Trav15-1-dv6-1
|
T cell receptor alpha variable 15-1-DV6-1 |
| chr5_+_92540444 | 2.25 |
ENSMUST00000126281.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr19_+_36325683 | 2.23 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr11_+_17161912 | 2.22 |
ENSMUST00000046955.7
|
Wdr92
|
WD repeat domain 92 |
| chr10_-_81186025 | 2.19 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr1_-_65225617 | 2.13 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr7_-_28661648 | 2.12 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr2_-_164699462 | 2.12 |
ENSMUST00000109316.8
ENSMUST00000156255.8 ENSMUST00000128110.2 ENSMUST00000109317.10 ENSMUST00000059954.14 |
Pltp
|
phospholipid transfer protein |
| chr10_-_81186137 | 2.11 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr17_+_32725420 | 2.09 |
ENSMUST00000235238.2
ENSMUST00000165999.2 |
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr2_-_155434487 | 2.07 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr9_+_107957621 | 2.03 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr6_-_131224305 | 1.99 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr9_+_72892850 | 1.99 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr5_-_116560916 | 1.99 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr5_-_66238313 | 1.98 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr4_+_116414855 | 1.93 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr11_+_55360502 | 1.85 |
ENSMUST00000018727.4
|
G3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr12_+_59142439 | 1.83 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_118367651 | 1.80 |
ENSMUST00000135383.9
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr5_-_38659449 | 1.73 |
ENSMUST00000005238.13
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr8_+_70735477 | 1.73 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr2_+_152360167 | 1.71 |
ENSMUST00000121912.2
|
Defb28
|
defensin beta 28 |
| chr11_+_73244561 | 1.66 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chr8_-_93806593 | 1.64 |
ENSMUST00000109582.3
|
Ces1b
|
carboxylesterase 1B |
| chr11_-_40646090 | 1.63 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr14_-_73563212 | 1.63 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr7_+_89780785 | 1.63 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr9_+_107957640 | 1.62 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr12_+_59176506 | 1.62 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr8_+_84748073 | 1.60 |
ENSMUST00000055077.7
|
Palm3
|
paralemmin 3 |
| chr9_-_110886576 | 1.58 |
ENSMUST00000199839.5
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chrX_+_73298342 | 1.56 |
ENSMUST00000096424.11
|
Emd
|
emerin |
| chr5_+_52521133 | 1.56 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr15_-_102259158 | 1.56 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr7_+_141047416 | 1.53 |
ENSMUST00000209988.2
|
Cd151
|
CD151 antigen |
| chr8_-_69541852 | 1.53 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr12_-_114330574 | 1.52 |
ENSMUST00000103485.3
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr1_+_78794475 | 1.49 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr9_+_32305259 | 1.48 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr9_+_5298669 | 1.46 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr17_-_35081129 | 1.45 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr6_-_29212295 | 1.44 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chrX_+_73298285 | 1.41 |
ENSMUST00000002029.13
|
Emd
|
emerin |
| chr12_-_113236868 | 1.41 |
ENSMUST00000223335.2
ENSMUST00000137336.3 |
Ighe
|
Immunoglobulin heavy constant epsilon |
| chr6_+_68547717 | 1.39 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr8_-_65471175 | 1.39 |
ENSMUST00000078409.5
ENSMUST00000048565.9 |
Trim61
Trim60
|
tripartite motif-containing 61 tripartite motif-containing 60 |
| chr4_+_155875629 | 1.38 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr7_-_3828640 | 1.37 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr3_+_20043315 | 1.35 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chr12_+_59176543 | 1.33 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr17_+_8463886 | 1.31 |
ENSMUST00000231545.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr1_-_131441962 | 1.30 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chrX_-_100777806 | 1.26 |
ENSMUST00000056614.7
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr16_+_20536415 | 1.21 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_-_65225572 | 1.20 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr4_-_135112956 | 1.19 |
ENSMUST00000105857.8
ENSMUST00000105858.8 ENSMUST00000064481.15 ENSMUST00000123632.2 |
Ncmap
|
noncompact myelin associated protein |
| chr9_+_72892693 | 1.19 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr9_+_107454114 | 1.19 |
ENSMUST00000112387.9
ENSMUST00000123005.8 ENSMUST00000010195.14 ENSMUST00000144392.2 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr14_+_53505374 | 1.18 |
ENSMUST00000199112.2
|
Trav15n-2
|
T cell receptor alpha variable 15N-2 |
| chr18_+_80296508 | 1.18 |
ENSMUST00000157056.8
|
Slc66a2
|
solute carrier family 66 member 2 |
| chr5_-_38637474 | 1.17 |
ENSMUST00000143758.8
ENSMUST00000156272.8 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr19_-_47126719 | 1.16 |
ENSMUST00000140512.8
ENSMUST00000035822.2 |
Calhm2
|
calcium homeostasis modulator family member 2 |
| chr2_+_163662752 | 1.16 |
ENSMUST00000029188.8
|
Ccn5
|
cellular communication network factor 5 |
| chr11_+_66802807 | 1.14 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr7_+_16043502 | 1.04 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr9_-_75448979 | 1.02 |
ENSMUST00000214171.2
|
Tmod3
|
tropomodulin 3 |
| chr1_+_174218612 | 1.02 |
ENSMUST00000075329.3
|
Olfr248
|
olfactory receptor 248 |
| chr3_+_29136172 | 1.01 |
ENSMUST00000124809.8
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr8_+_10027707 | 1.01 |
ENSMUST00000139793.8
ENSMUST00000048216.6 |
Abhd13
|
abhydrolase domain containing 13 |
| chr9_-_72892617 | 1.00 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr9_+_72892786 | 0.97 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr14_+_53411782 | 0.97 |
ENSMUST00000197433.5
ENSMUST00000103590.4 |
Trav15n-1
|
T cell receptor alpha variable 15N-1 |
| chr16_+_20536545 | 0.97 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_+_75879689 | 0.96 |
ENSMUST00000238633.2
ENSMUST00000022579.2 |
Erich6b
|
glutamate rich 6B |
| chr14_-_49303826 | 0.95 |
ENSMUST00000161504.8
|
Exoc5
|
exocyst complex component 5 |
| chr18_+_80296488 | 0.93 |
ENSMUST00000131780.8
|
Slc66a2
|
solute carrier family 66 member 2 |
| chr11_-_106192627 | 0.93 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr5_-_38659422 | 0.90 |
ENSMUST00000147664.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 23.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 2.6 | 10.4 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 2.4 | 9.7 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 2.2 | 6.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 2.0 | 10.1 | GO:2000721 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 2.0 | 5.9 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 2.0 | 5.9 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 1.9 | 7.8 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 1.8 | 7.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 1.5 | 7.4 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 1.4 | 4.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.3 | 5.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 1.3 | 3.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.3 | 5.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 1.3 | 11.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 1.1 | 4.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 1.0 | 8.3 | GO:0070268 | cornification(GO:0070268) |
| 1.0 | 6.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 1.0 | 4.8 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 1.0 | 5.7 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.9 | 5.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 2.7 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.8 | 4.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.8 | 3.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.8 | 6.5 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.8 | 4.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.7 | 2.8 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.7 | 4.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.7 | 5.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.6 | 4.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.6 | 3.7 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.6 | 2.8 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.6 | 4.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.5 | 3.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.5 | 2.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.5 | 4.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 6.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.4 | 1.3 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.4 | 9.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.4 | 11.7 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.4 | 7.8 | GO:2000675 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.4 | 2.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.4 | 1.2 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.4 | 6.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 1.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 5.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.4 | 10.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.3 | 2.7 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 2.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 5.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 5.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.3 | 1.6 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 5.9 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.3 | 0.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 1.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 9.1 | GO:0060445 | epithelial tube branching involved in lung morphogenesis(GO:0060441) branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 3.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 4.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 7.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 1.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.8 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.2 | 1.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 1.5 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 3.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 6.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 4.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.7 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.2 | 1.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 2.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.3 | GO:0021815 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.1 | 11.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 18.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 2.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 6.2 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 4.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 4.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.2 | GO:0046491 | L-methylmalonyl-CoA metabolic process(GO:0046491) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 5.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 3.7 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 2.1 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 6.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.9 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.2 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 8.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 4.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 3.4 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 1.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 5.4 | GO:0010324 | membrane invagination(GO:0010324) |
| 0.0 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 1.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 2.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.8 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.0 | GO:0060156 | positive regulation of glomerular filtration(GO:0003104) vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.0 | 5.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 4.7 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.5 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.0 | 1.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.9 | GO:0002687 | positive regulation of leukocyte migration(GO:0002687) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 20.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.2 | 6.6 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.1 | 10.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.1 | 5.6 | GO:0005914 | spot adherens junction(GO:0005914) |
| 1.1 | 4.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.6 | 4.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.6 | 2.4 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.5 | 6.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 5.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.5 | 13.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 1.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 5.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.3 | 4.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 2.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 5.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 14.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 10.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 0.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 14.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 7.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 8.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 8.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 7.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 1.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 5.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 7.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 12.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 20.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 1.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 6.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 2.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 6.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 41.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 5.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 9.5 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 6.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 35.9 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 23.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.6 | 7.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 2.0 | 8.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 2.0 | 5.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.7 | 8.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.5 | 4.6 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 1.5 | 9.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.2 | 9.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.1 | 5.7 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 1.1 | 7.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.1 | 5.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.9 | 6.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.9 | 3.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.8 | 3.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.7 | 2.8 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.6 | 6.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.6 | 8.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.6 | 7.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 6.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 6.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 3.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.5 | 9.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 4.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 4.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 2.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.4 | 1.5 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 5.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 2.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.3 | 2.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.3 | 3.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 1.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 1.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.3 | 0.9 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 5.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 1.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 8.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 10.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 4.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 2.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 7.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 6.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 0.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 4.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 4.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 2.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 0.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 2.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 2.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 2.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 4.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 3.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 4.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 6.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 5.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 6.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.1 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 11.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 7.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.2 | GO:0004493 | methylmalonyl-CoA epimerase activity(GO:0004493) |
| 0.1 | 1.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.7 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 2.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 14.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 9.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 7.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 7.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 2.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 13.2 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 12.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 3.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 50.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 7.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 9.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 21.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 10.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 2.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 15.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 12.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 7.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 3.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 6.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 5.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 5.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 7.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 1.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 3.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 0.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 7.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 7.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 11.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 4.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 4.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 20.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 2.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 2.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 9.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 5.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 30.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 4.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 4.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |