Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Trp73
Z-value: 0.60
Transcription factors associated with Trp73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp73
|
ENSMUSG00000029026.18 | Trp73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp73 | mm39_v1_chr4_-_154181562_154181630 | -0.21 | 8.0e-02 | Click! |
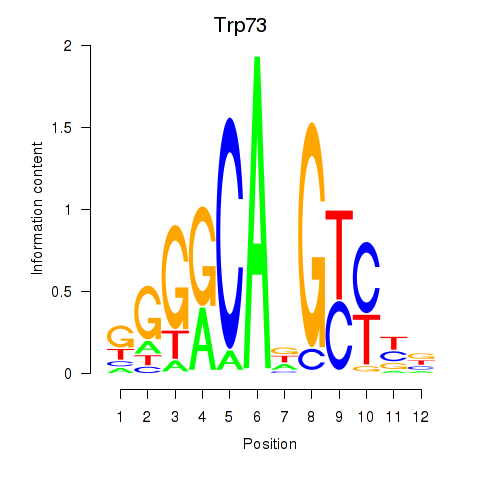
Activity profile of Trp73 motif
Sorted Z-values of Trp73 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp73
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_30487322 | 3.50 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr5_+_64960705 | 3.12 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr11_-_69838971 | 2.92 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr7_-_19530714 | 2.80 |
ENSMUST00000108449.9
ENSMUST00000043822.8 |
Cblc
|
Casitas B-lineage lymphoma c |
| chr11_-_103235475 | 2.73 |
ENSMUST00000041385.14
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr2_+_119068012 | 2.56 |
ENSMUST00000110817.3
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr2_+_119067832 | 2.53 |
ENSMUST00000028783.14
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr2_+_119067929 | 2.52 |
ENSMUST00000110816.8
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr4_-_49451134 | 2.44 |
ENSMUST00000107697.2
ENSMUST00000095086.3 |
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr3_+_159545309 | 2.37 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr8_-_71112295 | 2.27 |
ENSMUST00000211715.2
ENSMUST00000210307.2 ENSMUST00000209768.2 ENSMUST00000070173.9 |
Pgpep1
|
pyroglutamyl-peptidase I |
| chr2_-_127383305 | 2.17 |
ENSMUST00000103214.3
|
Prom2
|
prominin 2 |
| chr5_-_103777145 | 2.16 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr2_-_127383332 | 2.11 |
ENSMUST00000028855.14
|
Prom2
|
prominin 2 |
| chr17_+_28451674 | 2.11 |
ENSMUST00000002320.16
ENSMUST00000232879.2 ENSMUST00000166744.8 |
Ppard
|
peroxisome proliferator activator receptor delta |
| chr17_+_23883974 | 2.10 |
ENSMUST00000233541.2
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr15_+_98953044 | 1.82 |
ENSMUST00000230021.2
ENSMUST00000047104.15 ENSMUST00000024249.5 |
Prph
|
peripherin |
| chr18_-_32170012 | 1.81 |
ENSMUST00000134663.2
|
Myo7b
|
myosin VIIB |
| chr14_+_33807935 | 1.79 |
ENSMUST00000022519.15
|
Anxa8
|
annexin A8 |
| chr14_+_33807964 | 1.76 |
ENSMUST00000120077.2
|
Anxa8
|
annexin A8 |
| chr4_-_106662195 | 1.75 |
ENSMUST00000065253.13
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr14_+_70793340 | 1.74 |
ENSMUST00000163060.2
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr10_+_127478844 | 1.72 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr2_+_74522258 | 1.66 |
ENSMUST00000061745.5
|
Hoxd10
|
homeobox D10 |
| chr15_-_98505508 | 1.58 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr1_+_134217727 | 1.54 |
ENSMUST00000027730.6
|
Myog
|
myogenin |
| chr2_-_164699462 | 1.53 |
ENSMUST00000109316.8
ENSMUST00000156255.8 ENSMUST00000128110.2 ENSMUST00000109317.10 ENSMUST00000059954.14 |
Pltp
|
phospholipid transfer protein |
| chr11_+_32246489 | 1.46 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr10_+_69761597 | 1.38 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr5_+_137286535 | 1.36 |
ENSMUST00000024099.11
ENSMUST00000196208.5 ENSMUST00000085934.4 |
Ache
|
acetylcholinesterase |
| chr19_-_34724689 | 1.35 |
ENSMUST00000009522.4
|
Slc16a12
|
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
| chr7_+_26819334 | 1.34 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr4_-_87724533 | 1.31 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr4_-_87724512 | 1.30 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_-_29212239 | 1.25 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr10_+_75242745 | 1.25 |
ENSMUST00000039925.8
|
Upb1
|
ureidopropionase, beta |
| chr2_-_90735171 | 1.24 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr9_-_58065800 | 1.20 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr11_-_76359222 | 1.18 |
ENSMUST00000065028.14
|
Abr
|
active BCR-related gene |
| chr1_-_136188611 | 1.16 |
ENSMUST00000086395.7
|
Gpr25
|
G protein-coupled receptor 25 |
| chr10_+_52267702 | 1.13 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr6_-_29212295 | 1.11 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr11_+_32233511 | 1.11 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr1_+_55127110 | 1.10 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr4_-_141125885 | 1.09 |
ENSMUST00000133676.3
ENSMUST00000042617.14 |
Clcnka
|
chloride channel, voltage-sensitive Ka |
| chr1_-_55127183 | 1.08 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr1_-_55127312 | 1.06 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr4_-_49383576 | 1.03 |
ENSMUST00000107698.8
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr10_+_69761630 | 0.99 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr3_+_28752050 | 0.98 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr11_-_89587671 | 0.98 |
ENSMUST00000128717.9
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr10_+_69761314 | 0.95 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr11_+_97917746 | 0.93 |
ENSMUST00000017548.7
|
Rpl19
|
ribosomal protein L19 |
| chr19_-_47146203 | 0.89 |
ENSMUST00000178630.2
|
Calhm3
|
calcium homeostasis modulator 3 |
| chr8_+_27575611 | 0.88 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr16_+_10297923 | 0.87 |
ENSMUST00000230395.2
|
Ciita
|
class II transactivator |
| chr3_+_89153258 | 0.87 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr17_+_28910714 | 0.85 |
ENSMUST00000233250.2
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr5_+_135698881 | 0.81 |
ENSMUST00000153500.8
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr7_+_140462343 | 0.79 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr12_-_45120895 | 0.79 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr2_-_91854844 | 0.79 |
ENSMUST00000028663.5
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr17_+_29309942 | 0.78 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr16_+_10298011 | 0.76 |
ENSMUST00000230450.2
|
Ciita
|
class II transactivator |
| chr5_-_31117617 | 0.75 |
ENSMUST00000202567.2
ENSMUST00000074840.12 |
Preb
|
prolactin regulatory element binding |
| chr19_+_46293160 | 0.75 |
ENSMUST00000073116.13
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr7_+_140461860 | 0.74 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr9_+_108539296 | 0.74 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr9_-_44624496 | 0.73 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr10_-_67384898 | 0.72 |
ENSMUST00000075686.7
|
Ado
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr19_+_46293210 | 0.71 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr5_-_33940053 | 0.70 |
ENSMUST00000148451.2
ENSMUST00000005431.6 |
Letm1
|
leucine zipper-EF-hand containing transmembrane protein 1 |
| chr4_+_32238712 | 0.69 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr9_+_121245036 | 0.68 |
ENSMUST00000211187.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chrX_-_108056995 | 0.67 |
ENSMUST00000033597.9
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr11_-_33528933 | 0.66 |
ENSMUST00000020366.8
ENSMUST00000135350.2 |
Gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr2_-_170339061 | 0.65 |
ENSMUST00000038824.6
|
Cyp24a1
|
cytochrome P450, family 24, subfamily a, polypeptide 1 |
| chr9_-_32454157 | 0.65 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr18_-_36201664 | 0.60 |
ENSMUST00000239409.2
|
Nrg2
|
neuregulin 2 |
| chr11_-_95037089 | 0.56 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr7_-_4517367 | 0.56 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr4_+_119396913 | 0.55 |
ENSMUST00000137560.8
|
Foxj3
|
forkhead box J3 |
| chr16_+_4964849 | 0.52 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chr4_+_100926170 | 0.52 |
ENSMUST00000106955.2
ENSMUST00000038463.15 |
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr2_+_160573178 | 0.50 |
ENSMUST00000103115.8
|
Plcg1
|
phospholipase C, gamma 1 |
| chr11_+_60030891 | 0.50 |
ENSMUST00000171108.8
ENSMUST00000090806.5 |
Rai1
|
retinoic acid induced 1 |
| chr1_-_170042947 | 0.49 |
ENSMUST00000027979.14
ENSMUST00000123399.2 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr14_+_53607470 | 0.47 |
ENSMUST00000103652.5
|
Trav14n-3
|
T cell receptor alpha variable 14N-3 |
| chr6_+_40548291 | 0.46 |
ENSMUST00000051540.5
|
Olfr460
|
olfactory receptor 460 |
| chr19_-_7460550 | 0.45 |
ENSMUST00000088169.7
|
Rtn3
|
reticulon 3 |
| chr2_+_160573604 | 0.44 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr10_-_57408585 | 0.42 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr7_+_120411588 | 0.42 |
ENSMUST00000208668.2
|
Gm5737
|
predicted gene 5737 |
| chr11_-_99869879 | 0.41 |
ENSMUST00000105051.2
|
Krtap29-1
|
keratin associated protein 29-1 |
| chr10_+_115854118 | 0.40 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr18_+_56565188 | 0.38 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr6_-_97464761 | 0.37 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B |
| chr11_+_69914746 | 0.37 |
ENSMUST00000124568.10
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr2_+_127112127 | 0.34 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr19_-_7460600 | 0.34 |
ENSMUST00000235593.2
ENSMUST00000088171.12 ENSMUST00000065304.13 ENSMUST00000025667.14 |
Rtn3
|
reticulon 3 |
| chr7_-_4517608 | 0.33 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr1_+_167445815 | 0.32 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr19_+_41900360 | 0.30 |
ENSMUST00000011896.8
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr10_-_62486948 | 0.29 |
ENSMUST00000020270.6
|
Ddx50
|
DExD box helicase 50 |
| chr10_+_105676925 | 0.27 |
ENSMUST00000020049.9
|
Ccdc59
|
coiled-coil domain containing 59 |
| chr5_-_139805661 | 0.26 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr6_-_97408367 | 0.25 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chrX_+_102365004 | 0.25 |
ENSMUST00000033689.3
|
Cdx4
|
caudal type homeobox 4 |
| chr11_+_75546989 | 0.23 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr6_+_41095752 | 0.22 |
ENSMUST00000103269.3
|
Trbv12-2
|
T cell receptor beta, variable 12-2 |
| chr9_-_42383494 | 0.22 |
ENSMUST00000128959.8
ENSMUST00000066148.12 ENSMUST00000138506.8 |
Tbcel
|
tubulin folding cofactor E-like |
| chr10_-_57408512 | 0.17 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr7_-_4517559 | 0.14 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr17_-_46513499 | 0.14 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr9_+_101815680 | 0.13 |
ENSMUST00000112841.2
|
9630041A04Rik
|
RIKEN cDNA 9630041A04 gene |
| chr10_+_110581293 | 0.11 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr16_+_35891991 | 0.10 |
ENSMUST00000164916.9
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr7_-_141023199 | 0.10 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr14_-_52341426 | 0.08 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr2_-_114031897 | 0.07 |
ENSMUST00000050668.4
|
Zfp770
|
zinc finger protein 770 |
| chr12_+_111540920 | 0.06 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr19_+_46292759 | 0.06 |
ENSMUST00000237330.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr14_+_52818032 | 0.06 |
ENSMUST00000103569.3
|
Trav3-1
|
T cell receptor alpha variable 3-1 |
| chr19_+_53317844 | 0.05 |
ENSMUST00000111737.3
ENSMUST00000025998.15 ENSMUST00000237837.2 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr14_-_24295988 | 0.04 |
ENSMUST00000073687.13
ENSMUST00000090398.11 |
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr16_-_56748424 | 0.04 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr3_+_53396120 | 0.03 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr12_+_3415107 | 0.03 |
ENSMUST00000220210.2
|
Kif3c
|
kinesin family member 3C |
| chr2_+_59442378 | 0.02 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_+_185095232 | 0.00 |
ENSMUST00000046514.13
|
Eprs
|
glutamyl-prolyl-tRNA synthetase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.8 | 2.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.7 | 2.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 1.6 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.5 | 1.5 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 1.4 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.4 | 1.3 | GO:0015881 | creatine transport(GO:0015881) |
| 0.4 | 1.3 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 1.7 | GO:0035771 | T-helper 1 cell lineage commitment(GO:0002296) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.4 | 1.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.4 | 1.8 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.4 | 7.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 2.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 1.5 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 3.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.3 | 1.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 2.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.8 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 1.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 1.1 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.2 | 0.9 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.2 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 1.6 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.2 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.7 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 2.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 2.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.9 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.6 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 1.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.9 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 1.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.7 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 5.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 2.5 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.6 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.9 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 1.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.4 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 3.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0044393 | microspike(GO:0044393) |
| 0.5 | 2.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 2.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 0.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 3.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 3.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.7 | 2.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.6 | 2.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.5 | 1.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 1.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.4 | 2.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 2.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.3 | 0.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 0.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.8 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 5.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 2.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 2.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.7 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 1.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 4.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 7.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 2.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0035326 | enhancer binding(GO:0035326) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.3 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |