Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Twist1
Z-value: 0.93
Transcription factors associated with Twist1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Twist1
|
ENSMUSG00000035799.7 | Twist1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | mm39_v1_chr12_+_34007645_34007670 | 0.30 | 1.1e-02 | Click! |
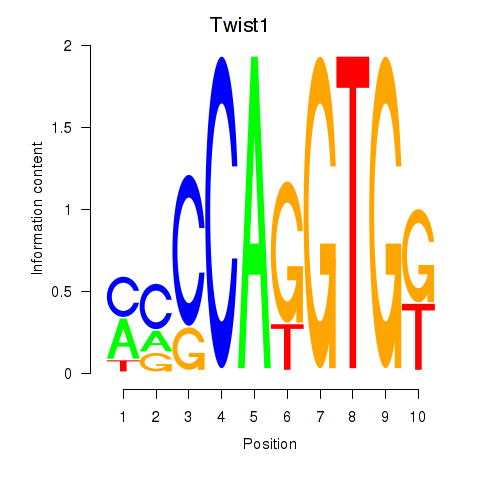
Activity profile of Twist1 motif
Sorted Z-values of Twist1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Twist1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41012435 | 5.51 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr6_+_30639217 | 4.84 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr17_+_34029484 | 4.49 |
ENSMUST00000048560.11
ENSMUST00000172649.8 ENSMUST00000173789.2 |
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr2_+_71811526 | 4.04 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_46203047 | 3.72 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr9_-_54569128 | 3.52 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr8_+_55024446 | 3.43 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr19_-_11243530 | 3.30 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr5_+_8943943 | 3.22 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr4_+_155553571 | 3.21 |
ENSMUST00000178188.8
ENSMUST00000178238.8 ENSMUST00000023920.3 |
Tmem52
|
transmembrane protein 52 |
| chr8_-_106660470 | 3.11 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr8_+_93084253 | 2.58 |
ENSMUST00000210246.2
ENSMUST00000034184.12 |
Irx5
|
Iroquois homeobox 5 |
| chr5_+_24569802 | 2.54 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr19_+_5790918 | 2.41 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chrX_-_56371953 | 2.36 |
ENSMUST00000176986.8
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr17_+_75772475 | 2.34 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr7_-_44174065 | 2.33 |
ENSMUST00000165208.4
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr9_-_54568950 | 2.30 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr11_-_113600346 | 2.25 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr10_+_70080913 | 2.23 |
ENSMUST00000046807.7
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr11_+_109541400 | 2.22 |
ENSMUST00000106676.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr8_-_78337297 | 2.16 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr4_-_152122891 | 2.10 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr13_-_58261406 | 2.09 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr4_-_123458465 | 2.03 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr15_-_100497863 | 2.01 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chrX_-_100310959 | 2.00 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr9_-_14815228 | 1.99 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr16_-_59421342 | 1.89 |
ENSMUST00000172910.3
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr2_+_31777273 | 1.81 |
ENSMUST00000138325.8
ENSMUST00000028187.7 |
Lamc3
|
laminin gamma 3 |
| chr18_-_16942289 | 1.74 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr9_+_65172455 | 1.69 |
ENSMUST00000048762.8
|
Cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr18_+_61044830 | 1.62 |
ENSMUST00000040359.6
|
Arsi
|
arylsulfatase i |
| chr5_-_23880939 | 1.62 |
ENSMUST00000196388.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr4_-_152561896 | 1.61 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr4_-_59915035 | 1.58 |
ENSMUST00000030081.2
|
Slc46a2
|
solute carrier family 46, member 2 |
| chr9_-_14815050 | 1.56 |
ENSMUST00000148155.2
ENSMUST00000121116.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr7_-_126817639 | 1.53 |
ENSMUST00000152267.8
ENSMUST00000106314.8 |
Septin1
|
septin 1 |
| chr7_+_45067747 | 1.52 |
ENSMUST00000238638.2
|
Lhb
|
luteinizing hormone beta |
| chr7_-_30814652 | 1.50 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr5_-_35886605 | 1.48 |
ENSMUST00000070203.14
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr10_+_61010983 | 1.47 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr10_-_126866682 | 1.46 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr6_-_29164981 | 1.44 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr9_+_57037974 | 1.40 |
ENSMUST00000160147.8
ENSMUST00000161663.8 ENSMUST00000034836.16 ENSMUST00000161182.8 |
Man2c1
|
mannosidase, alpha, class 2C, member 1 |
| chr5_+_31454787 | 1.40 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr19_-_28941264 | 1.37 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr7_-_126817475 | 1.34 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr13_+_73775001 | 1.34 |
ENSMUST00000022104.9
|
Tert
|
telomerase reverse transcriptase |
| chr10_+_12936248 | 1.34 |
ENSMUST00000193426.6
|
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr6_-_124410452 | 1.30 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr8_+_107237483 | 1.30 |
ENSMUST00000080797.8
|
Cdh3
|
cadherin 3 |
| chr8_+_123148759 | 1.30 |
ENSMUST00000050963.4
|
Il17c
|
interleukin 17C |
| chr2_+_29509704 | 1.28 |
ENSMUST00000095087.11
ENSMUST00000091146.12 ENSMUST00000102872.11 ENSMUST00000147755.10 |
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr18_+_4920513 | 1.27 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr9_-_44253630 | 1.27 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr12_+_4642987 | 1.27 |
ENSMUST00000062580.8
ENSMUST00000220311.2 |
Itsn2
|
intersectin 2 |
| chr2_-_45007407 | 1.26 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_126866658 | 1.26 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr9_-_114673158 | 1.21 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr15_-_72932853 | 1.19 |
ENSMUST00000170633.9
ENSMUST00000228960.2 |
Trappc9
|
trafficking protein particle complex 9 |
| chr9_-_44253588 | 1.17 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr11_-_68817875 | 1.16 |
ENSMUST00000038932.14
ENSMUST00000125134.2 |
Odf4
|
outer dense fiber of sperm tails 4 |
| chr16_+_18630522 | 1.16 |
ENSMUST00000115578.10
|
Ufd1
|
ubiquitin recognition factor in ER-associated degradation 1 |
| chr11_-_113600838 | 1.15 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr15_-_89007290 | 1.14 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr11_+_87628356 | 1.14 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr2_-_65068917 | 1.13 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr5_+_105667254 | 1.10 |
ENSMUST00000067924.13
ENSMUST00000150981.2 |
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr2_+_139520098 | 1.09 |
ENSMUST00000184404.8
ENSMUST00000099307.4 |
Ism1
|
isthmin 1, angiogenesis inhibitor |
| chr13_+_55612050 | 1.08 |
ENSMUST00000046533.9
|
Prr7
|
proline rich 7 (synaptic) |
| chr7_+_30121776 | 1.08 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr11_+_53410552 | 1.06 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr5_-_23881353 | 1.04 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr4_-_129452180 | 1.03 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_129452148 | 1.03 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr5_+_149335214 | 1.02 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr8_+_72889073 | 1.02 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr11_-_94440025 | 1.01 |
ENSMUST00000040487.4
|
Rsad1
|
radical S-adenosyl methionine domain containing 1 |
| chr9_-_108444561 | 1.00 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr17_+_34258411 | 0.97 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr2_+_163867370 | 0.96 |
ENSMUST00000067715.5
|
Pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr6_-_83808717 | 0.91 |
ENSMUST00000058383.9
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr8_+_122996308 | 0.91 |
ENSMUST00000055537.3
|
Zfp469
|
zinc finger protein 469 |
| chr11_+_109541747 | 0.91 |
ENSMUST00000049527.7
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr7_-_139616538 | 0.91 |
ENSMUST00000211638.2
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr7_+_82516491 | 0.90 |
ENSMUST00000082237.7
|
Mex3b
|
mex3 RNA binding family member B |
| chr7_+_130179063 | 0.89 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chrX_-_18327610 | 0.88 |
ENSMUST00000044188.5
|
Dipk2b
|
divergent protein kinase domain 2B |
| chr7_-_140596811 | 0.88 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr14_-_70414236 | 0.87 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr4_+_155875629 | 0.87 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr13_+_97208069 | 0.85 |
ENSMUST00000042517.8
|
Fam169a
|
family with sequence similarity 169, member A |
| chr8_+_53964721 | 0.82 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr7_+_139616298 | 0.81 |
ENSMUST00000168194.3
ENSMUST00000210882.2 |
Zfp511
|
zinc finger protein 511 |
| chr9_-_14815163 | 0.77 |
ENSMUST00000069408.10
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr2_+_155593030 | 0.73 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chrX_+_56098908 | 0.67 |
ENSMUST00000114751.9
ENSMUST00000088652.6 |
Htatsf1
|
HIV TAT specific factor 1 |
| chr2_+_127183838 | 0.66 |
ENSMUST00000156747.2
|
Astl
|
astacin-like metalloendopeptidase (M12 family) |
| chr7_-_84339156 | 0.65 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr7_-_84339045 | 0.64 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr17_-_35978438 | 0.64 |
ENSMUST00000043674.15
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr5_+_31454939 | 0.60 |
ENSMUST00000201675.3
|
Gckr
|
glucokinase regulatory protein |
| chrX_+_85235370 | 0.60 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr16_+_18245134 | 0.60 |
ENSMUST00000205679.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr16_+_31482745 | 0.57 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr14_+_101891416 | 0.57 |
ENSMUST00000002289.8
|
Uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr3_+_107008343 | 0.54 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_142743438 | 0.53 |
ENSMUST00000230763.2
ENSMUST00000043589.8 |
Kif16b
|
kinesin family member 16B |
| chr15_+_100202642 | 0.52 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr4_-_126362372 | 0.51 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr2_-_180562718 | 0.47 |
ENSMUST00000037299.15
ENSMUST00000108876.9 |
Ythdf1
|
YTH N6-methyladenosine RNA binding protein 1 |
| chr2_+_173952123 | 0.39 |
ENSMUST00000044415.16
|
Npepl1
|
aminopeptidase-like 1 |
| chr2_+_130116357 | 0.38 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr3_+_89958940 | 0.37 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr2_-_32674435 | 0.36 |
ENSMUST00000102813.2
|
Cfap157
|
cilia and flagella associated protein 157 |
| chr11_-_109188947 | 0.35 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr14_-_70855980 | 0.34 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr10_-_23226034 | 0.34 |
ENSMUST00000219315.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr7_+_24310171 | 0.34 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr16_+_31482658 | 0.32 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr17_+_8559539 | 0.32 |
ENSMUST00000163887.2
|
Prr18
|
proline rich 18 |
| chr3_+_65435825 | 0.30 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr9_-_44920899 | 0.30 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr7_-_80051455 | 0.29 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_-_172782089 | 0.29 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr1_+_90926443 | 0.28 |
ENSMUST00000189505.7
ENSMUST00000185531.7 ENSMUST00000068116.13 |
Lrrfip1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr1_+_174329361 | 0.27 |
ENSMUST00000030039.13
|
Fmn2
|
formin 2 |
| chr4_+_132495636 | 0.26 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr14_+_53061814 | 0.26 |
ENSMUST00000103648.4
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr8_-_117809188 | 0.24 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr19_-_24257904 | 0.24 |
ENSMUST00000081333.11
|
Fxn
|
frataxin |
| chr7_-_142233270 | 0.24 |
ENSMUST00000162317.2
ENSMUST00000125933.2 ENSMUST00000105931.8 ENSMUST00000105930.8 ENSMUST00000105933.8 ENSMUST00000105932.2 ENSMUST00000000220.3 |
Ins2
|
insulin II |
| chr11_-_109188917 | 0.24 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr7_+_127400016 | 0.22 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_-_104869237 | 0.16 |
ENSMUST00000029429.6
|
Wnt2b
|
wingless-type MMTV integration site family, member 2B |
| chr14_+_58313964 | 0.15 |
ENSMUST00000166770.2
|
Fgf9
|
fibroblast growth factor 9 |
| chr2_-_168607166 | 0.13 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr6_-_119940694 | 0.13 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr17_+_74645936 | 0.12 |
ENSMUST00000224711.2
ENSMUST00000024869.8 ENSMUST00000233611.2 |
Spast
|
spastin |
| chr8_-_32499513 | 0.11 |
ENSMUST00000208205.3
|
Nrg1
|
neuregulin 1 |
| chr11_-_82761954 | 0.09 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr11_-_109188892 | 0.09 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr2_+_29855572 | 0.08 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr11_+_69826603 | 0.07 |
ENSMUST00000018698.12
|
Ybx2
|
Y box protein 2 |
| chr9_+_118307918 | 0.07 |
ENSMUST00000150633.2
|
Eomes
|
eomesodermin |
| chr10_+_69369854 | 0.06 |
ENSMUST00000182557.8
|
Ank3
|
ankyrin 3, epithelial |
| chr15_-_57755753 | 0.05 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr11_+_69826719 | 0.04 |
ENSMUST00000149194.8
|
Ybx2
|
Y box protein 2 |
| chr13_+_104246259 | 0.03 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr18_+_40390013 | 0.02 |
ENSMUST00000096572.2
ENSMUST00000236889.2 |
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr3_-_141874955 | 0.00 |
ENSMUST00000098568.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.8 | 2.5 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.8 | 2.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.8 | 4.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.8 | 2.4 | GO:1902462 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 2.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 1.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.4 | 1.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.4 | 2.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 1.3 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.4 | 3.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 2.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.3 | 2.0 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.3 | 3.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.0 | GO:0009750 | response to fructose(GO:0009750) |
| 0.3 | 2.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.7 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 4.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 0.9 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 2.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 3.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 1.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 2.0 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 2.1 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 2.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 2.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:2000360 | prevention of polyspermy(GO:0060468) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 1.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 2.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.6 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 5.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 1.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:1900104 | neural fold elevation formation(GO:0021502) hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 1.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 2.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.5 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 4.5 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 1.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.2 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 1.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.5 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.2 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 0.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.9 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 1.6 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 1.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.9 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 1.0 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 1.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.5 | 1.6 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.4 | 1.2 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.2 | 0.9 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.7 | GO:0060473 | cortical granule(GO:0060473) |
| 0.2 | 3.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 3.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 3.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.3 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 1.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 5.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 7.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.6 | 2.5 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.5 | 3.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.4 | 2.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 2.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.4 | 5.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 3.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 0.7 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.2 | 2.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 4.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 4.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 1.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 2.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 2.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 4.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 1.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.0 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.2 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.1 | 2.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 3.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 8.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 6.0 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 5.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 3.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 4.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 3.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.3 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 3.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |