Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
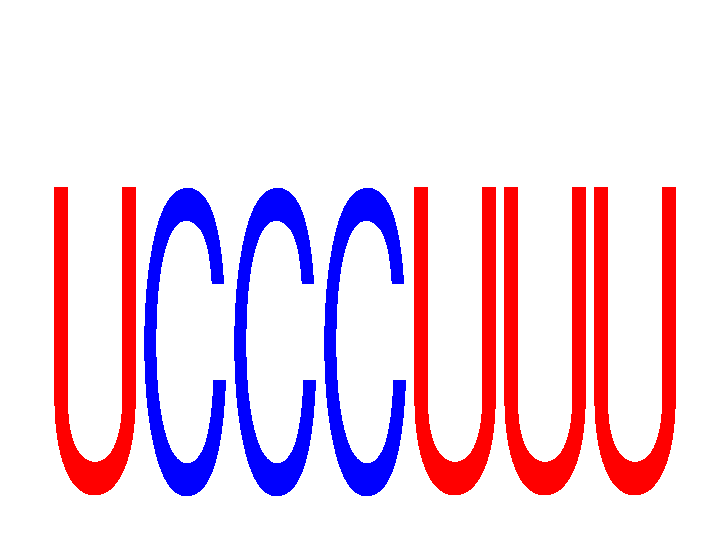
Results for UCCCUUU
Z-value: 0.40
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-204-5p
|
MIMAT0000237 |
|
mmu-miR-211-5p
|
MIMAT0000668 |
|
mmu-miR-7670-3p
|
MIMAT0029847 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_55407872 | 2.69 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr13_+_45660905 | 1.59 |
ENSMUST00000000260.13
|
Gmpr
|
guanosine monophosphate reductase |
| chr13_+_74157538 | 1.58 |
ENSMUST00000022057.9
|
Tppp
|
tubulin polymerization promoting protein |
| chr4_+_101276474 | 1.56 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr18_-_16942289 | 1.53 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr9_-_21963306 | 1.39 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr15_+_80057894 | 1.36 |
ENSMUST00000044970.7
|
Mgat3
|
mannoside acetylglucosaminyltransferase 3 |
| chr15_-_78004211 | 1.30 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr11_-_69451012 | 1.25 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr14_+_121272950 | 1.22 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr5_-_51711237 | 1.18 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr6_-_113320858 | 1.10 |
ENSMUST00000155543.2
ENSMUST00000032409.15 |
Camk1
|
calcium/calmodulin-dependent protein kinase I |
| chr1_+_75522902 | 1.09 |
ENSMUST00000124341.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr11_+_17109263 | 1.08 |
ENSMUST00000102880.5
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
| chr6_-_57512355 | 1.08 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chrX_+_92330102 | 1.01 |
ENSMUST00000046565.13
ENSMUST00000113947.6 |
Arx
|
aristaless related homeobox |
| chr7_+_44813363 | 1.00 |
ENSMUST00000085374.7
ENSMUST00000209634.2 |
Slc17a7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 |
| chr9_+_57428490 | 0.94 |
ENSMUST00000000090.8
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr15_+_58805605 | 0.91 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr17_+_81251997 | 0.90 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr7_+_57069417 | 0.83 |
ENSMUST00000085240.11
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr7_+_141142075 | 0.83 |
ENSMUST00000003038.12
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr1_+_187730018 | 0.82 |
ENSMUST00000027906.13
|
Esrrg
|
estrogen-related receptor gamma |
| chr5_+_96521814 | 0.80 |
ENSMUST00000036019.5
|
Fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr13_+_41013230 | 0.79 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_-_73722874 | 0.78 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chr16_+_41353360 | 0.76 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr6_-_39354570 | 0.76 |
ENSMUST00000200771.4
ENSMUST00000202204.4 ENSMUST00000202952.2 ENSMUST00000202400.4 ENSMUST00000200969.4 ENSMUST00000202749.4 ENSMUST00000090243.8 |
Slc37a3
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 3 |
| chr4_+_102617495 | 0.75 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr7_+_4122523 | 0.75 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr10_-_9550907 | 0.74 |
ENSMUST00000100070.5
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr14_-_39194782 | 0.72 |
ENSMUST00000168810.9
ENSMUST00000173780.2 ENSMUST00000166968.9 |
Nrg3
|
neuregulin 3 |
| chr4_+_149602673 | 0.72 |
ENSMUST00000030839.13
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr18_+_77032080 | 0.68 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr2_+_107120934 | 0.64 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chrX_-_143471176 | 0.64 |
ENSMUST00000040184.4
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr4_-_46991842 | 0.62 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr8_+_77626400 | 0.61 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr7_-_105049197 | 0.61 |
ENSMUST00000048079.14
ENSMUST00000118726.8 ENSMUST00000074686.10 ENSMUST00000122327.7 ENSMUST00000179474.9 |
Fam160a2
|
family with sequence similarity 160, member A2 |
| chr4_+_152423075 | 0.60 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr6_+_17749169 | 0.60 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr15_+_39061612 | 0.59 |
ENSMUST00000082054.12
ENSMUST00000227243.2 ENSMUST00000042917.10 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr11_-_88742285 | 0.59 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr14_-_24295988 | 0.57 |
ENSMUST00000073687.13
ENSMUST00000090398.11 |
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr14_-_25769457 | 0.56 |
ENSMUST00000069180.8
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr4_+_57434247 | 0.53 |
ENSMUST00000102905.8
|
Pakap
|
paralemmin A kinase anchor protein |
| chr10_-_39901249 | 0.49 |
ENSMUST00000163705.3
|
Mfsd4b1
|
major facilitator superfamily domain containing 4B1 |
| chrX_-_63320543 | 0.49 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr6_-_92191878 | 0.49 |
ENSMUST00000014694.11
|
Rbsn
|
rabenosyn, RAB effector |
| chr13_+_31990604 | 0.49 |
ENSMUST00000062292.5
|
Foxc1
|
forkhead box C1 |
| chr10_+_126914755 | 0.49 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr3_+_62245765 | 0.48 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr18_-_72484126 | 0.47 |
ENSMUST00000114943.11
|
Dcc
|
deleted in colorectal carcinoma |
| chr3_+_129326004 | 0.47 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr12_-_46865709 | 0.45 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr2_-_130681978 | 0.45 |
ENSMUST00000044766.15
ENSMUST00000138990.2 ENSMUST00000120316.8 ENSMUST00000110243.8 |
4930402H24Rik
|
RIKEN cDNA 4930402H24 gene |
| chr1_+_153528689 | 0.42 |
ENSMUST00000041776.12
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_+_28813125 | 0.41 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr1_-_190643922 | 0.38 |
ENSMUST00000159066.2
ENSMUST00000061611.15 |
Rps6kc1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr2_-_57004933 | 0.37 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr14_-_61794330 | 0.36 |
ENSMUST00000022497.15
|
Spryd7
|
SPRY domain containing 7 |
| chr9_-_60418286 | 0.35 |
ENSMUST00000098660.10
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr10_-_84276454 | 0.35 |
ENSMUST00000020220.15
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr8_-_124709859 | 0.35 |
ENSMUST00000075578.7
|
Abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr15_-_77191079 | 0.34 |
ENSMUST00000171751.10
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_-_87159237 | 0.34 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr13_+_41403317 | 0.33 |
ENSMUST00000165561.4
|
Smim13
|
small integral membrane protein 13 |
| chrX_+_92698469 | 0.33 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr6_+_41582416 | 0.32 |
ENSMUST00000114732.3
|
Ephb6
|
Eph receptor B6 |
| chrX_-_56549092 | 0.31 |
ENSMUST00000057645.6
|
Gpr101
|
G protein-coupled receptor 101 |
| chr2_+_116951855 | 0.30 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chrX_+_100892981 | 0.30 |
ENSMUST00000124279.6
ENSMUST00000101339.8 |
Nhsl2
|
NHS-like 2 |
| chr5_-_135963408 | 0.29 |
ENSMUST00000198270.2
ENSMUST00000055808.6 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| chr8_-_39128662 | 0.29 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr16_+_56942050 | 0.29 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr7_+_109721230 | 0.29 |
ENSMUST00000033326.10
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr1_-_84262274 | 0.28 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr15_+_68800261 | 0.27 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr2_-_5719302 | 0.27 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr2_+_151923449 | 0.26 |
ENSMUST00000064061.4
|
Scrt2
|
scratch family zinc finger 2 |
| chr18_+_23885390 | 0.24 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_179084220 | 0.24 |
ENSMUST00000000314.13
|
Cdh4
|
cadherin 4 |
| chr6_-_31540913 | 0.24 |
ENSMUST00000026698.8
|
Podxl
|
podocalyxin-like |
| chrX_-_23231245 | 0.24 |
ENSMUST00000115313.8
|
Klhl13
|
kelch-like 13 |
| chr8_+_13209141 | 0.24 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr2_+_109747984 | 0.23 |
ENSMUST00000046548.14
ENSMUST00000111037.3 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr19_-_18978463 | 0.23 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr1_-_34882131 | 0.22 |
ENSMUST00000167518.8
ENSMUST00000047534.12 |
Fam168b
|
family with sequence similarity 168, member B |
| chr5_-_123887434 | 0.22 |
ENSMUST00000182955.8
ENSMUST00000182489.8 ENSMUST00000183147.9 ENSMUST00000050827.14 ENSMUST00000057795.12 ENSMUST00000111515.8 ENSMUST00000182309.8 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr1_-_170695328 | 0.22 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr2_-_33777874 | 0.22 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr10_-_29411857 | 0.21 |
ENSMUST00000092623.5
|
Rspo3
|
R-spondin 3 |
| chr11_+_20151406 | 0.21 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr1_-_57011595 | 0.20 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr9_-_106764627 | 0.20 |
ENSMUST00000055843.9
|
Rbm15b
|
RNA binding motif protein 15B |
| chr12_-_3359924 | 0.20 |
ENSMUST00000021001.10
|
Rab10
|
RAB10, member RAS oncogene family |
| chr18_-_64794338 | 0.19 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr2_-_156833932 | 0.18 |
ENSMUST00000109558.2
ENSMUST00000069600.13 ENSMUST00000072298.13 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr2_+_25070749 | 0.18 |
ENSMUST00000104999.4
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr2_-_103627937 | 0.17 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chr13_-_38842967 | 0.17 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chrX_-_55643429 | 0.17 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr6_+_108760025 | 0.17 |
ENSMUST00000032196.9
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr11_-_120464062 | 0.17 |
ENSMUST00000026122.11
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr3_-_56091096 | 0.17 |
ENSMUST00000029374.8
|
Nbea
|
neurobeachin |
| chr15_-_98505508 | 0.14 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr12_-_104831266 | 0.14 |
ENSMUST00000109937.9
|
Clmn
|
calmin |
| chr8_-_91074971 | 0.14 |
ENSMUST00000109621.10
|
Tox3
|
TOX high mobility group box family member 3 |
| chr2_-_5900130 | 0.14 |
ENSMUST00000026926.5
ENSMUST00000193792.6 ENSMUST00000102981.10 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr3_+_95801263 | 0.14 |
ENSMUST00000015894.12
|
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr6_+_114620054 | 0.14 |
ENSMUST00000032457.17
|
Atg7
|
autophagy related 7 |
| chr19_+_5088854 | 0.13 |
ENSMUST00000053705.8
ENSMUST00000235776.2 |
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chr10_-_122912272 | 0.13 |
ENSMUST00000219203.2
ENSMUST00000073792.11 |
Mon2
|
MON2 homolog, regulator of endosome to Golgi trafficking |
| chr8_+_122317100 | 0.13 |
ENSMUST00000034270.17
ENSMUST00000181826.2 ENSMUST00000181948.2 |
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr9_+_103879745 | 0.12 |
ENSMUST00000035167.15
ENSMUST00000194774.6 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr12_+_83810402 | 0.11 |
ENSMUST00000121733.8
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chrX_+_162691978 | 0.11 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr13_-_45155616 | 0.11 |
ENSMUST00000072329.15
|
Dtnbp1
|
dystrobrevin binding protein 1 |
| chr8_+_26609384 | 0.10 |
ENSMUST00000014022.15
ENSMUST00000209300.2 ENSMUST00000153528.8 ENSMUST00000209707.2 |
Rnf170
|
ring finger protein 170 |
| chr18_+_51250748 | 0.10 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr11_-_76737794 | 0.10 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr8_-_88531018 | 0.10 |
ENSMUST00000165770.9
|
Zfp423
|
zinc finger protein 423 |
| chr11_+_72580823 | 0.10 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr18_-_39623698 | 0.10 |
ENSMUST00000115567.8
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_-_76091931 | 0.10 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr8_-_122379631 | 0.09 |
ENSMUST00000046386.5
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr1_-_43137768 | 0.09 |
ENSMUST00000190427.7
ENSMUST00000095014.8 |
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr8_+_110595216 | 0.08 |
ENSMUST00000179721.8
ENSMUST00000034175.5 |
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr5_+_105847811 | 0.08 |
ENSMUST00000060531.16
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr5_+_124577952 | 0.08 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr14_+_55120875 | 0.08 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr4_+_57845240 | 0.07 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr5_+_108609087 | 0.07 |
ENSMUST00000112597.8
ENSMUST00000046975.12 |
Pcgf3
|
polycomb group ring finger 3 |
| chr2_-_52314837 | 0.07 |
ENSMUST00000036541.8
|
Arl5a
|
ADP-ribosylation factor-like 5A |
| chr7_+_99184645 | 0.07 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr4_+_117109204 | 0.07 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr11_+_80099843 | 0.06 |
ENSMUST00000077451.14
ENSMUST00000055056.16 |
Rhot1
|
ras homolog family member T1 |
| chr2_+_31462780 | 0.06 |
ENSMUST00000137889.7
ENSMUST00000194386.6 ENSMUST00000055244.13 |
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr18_+_69478790 | 0.06 |
ENSMUST00000202116.4
ENSMUST00000114982.8 ENSMUST00000078486.13 ENSMUST00000202772.4 ENSMUST00000201288.4 |
Tcf4
|
transcription factor 4 |
| chr4_+_57637817 | 0.05 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr19_-_12773472 | 0.05 |
ENSMUST00000038627.9
|
Zfp91
|
zinc finger protein 91 |
| chr6_-_118174679 | 0.05 |
ENSMUST00000088790.9
|
Ret
|
ret proto-oncogene |
| chr4_-_58553311 | 0.04 |
ENSMUST00000107571.8
ENSMUST00000055018.11 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr6_-_138020409 | 0.04 |
ENSMUST00000111873.8
ENSMUST00000141280.3 |
Slc15a5
|
solute carrier family 15, member 5 |
| chr5_-_135280063 | 0.04 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr12_-_27392356 | 0.03 |
ENSMUST00000079063.7
|
Sox11
|
SRY (sex determining region Y)-box 11 |
| chr12_+_81073573 | 0.03 |
ENSMUST00000110347.9
ENSMUST00000021564.11 ENSMUST00000129362.2 |
Smoc1
|
SPARC related modular calcium binding 1 |
| chr5_+_110324506 | 0.01 |
ENSMUST00000112512.8
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr2_-_69416365 | 0.01 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr1_-_163141230 | 0.01 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr7_-_128063354 | 0.01 |
ENSMUST00000106226.9
ENSMUST00000106228.8 |
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr9_-_113537277 | 0.01 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr5_+_77122530 | 0.00 |
ENSMUST00000101087.10
ENSMUST00000120550.2 |
Srp72
|
signal recognition particle 72 |
| chr7_+_127344942 | 0.00 |
ENSMUST00000189562.7
ENSMUST00000186116.7 |
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1904633 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.3 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 1.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 0.6 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 0.8 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 | 1.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.6 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.1 | 0.6 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.1 | 1.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.5 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.3 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.1 | 0.2 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 1.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 1.6 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 0.4 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.6 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 2.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0061646 | positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization(GO:0061646) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 1.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.4 | 1.2 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.2 | 1.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.8 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.2 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.1 | 1.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.5 | 1.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.6 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 1.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 2.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |