Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for UGACCUA
Z-value: 0.16
miRNA associated with seed UGACCUA
| Name | miRBASE accession |
|---|---|
|
mmu-miR-192-5p
|
MIMAT0000517 |
|
mmu-miR-215-5p
|
MIMAT0000904 |
Activity profile of UGACCUA motif
Sorted Z-values of UGACCUA motif
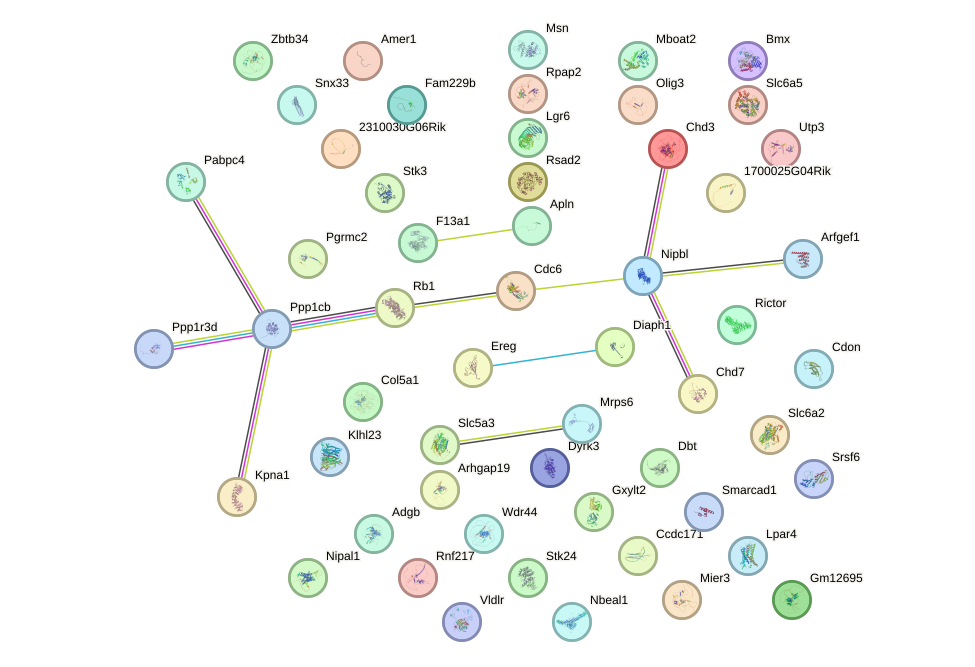
Network of associatons between targets according to the STRING database.
First level regulatory network of UGACCUA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_37234213 | 0.33 |
ENSMUST00000164727.8
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr12_-_26506422 | 0.29 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chrX_+_95139639 | 0.27 |
ENSMUST00000117399.2
|
Msn
|
moesin |
| chr4_+_123176570 | 0.25 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr10_+_87695117 | 0.24 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr13_+_111822712 | 0.21 |
ENSMUST00000109272.9
|
Mier3
|
MIER family member 3 |
| chr5_-_136596299 | 0.19 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr5_+_73808718 | 0.19 |
ENSMUST00000071077.14
|
Spata18
|
spermatogenesis associated 18 |
| chrX_-_163041185 | 0.18 |
ENSMUST00000112265.9
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chrX_+_105964224 | 0.17 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr2_+_27776428 | 0.16 |
ENSMUST00000028280.14
|
Col5a1
|
collagen, type V, alpha 1 |
| chr4_-_96673423 | 0.16 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr1_-_54965470 | 0.15 |
ENSMUST00000179030.8
ENSMUST00000044359.16 |
Ankrd44
|
ankyrin repeat domain 44 |
| chr19_+_27194757 | 0.15 |
ENSMUST00000047645.13
ENSMUST00000167487.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr3_-_27764571 | 0.15 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr4_+_8691303 | 0.14 |
ENSMUST00000051558.10
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr2_-_45003270 | 0.13 |
ENSMUST00000202935.4
ENSMUST00000068415.11 ENSMUST00000127520.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr9_-_56835633 | 0.13 |
ENSMUST00000050916.7
|
Snx33
|
sorting nexin 33 |
| chr19_-_41790458 | 0.13 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr11_+_98798627 | 0.13 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr6_+_100681670 | 0.13 |
ENSMUST00000032157.9
|
Gxylt2
|
glucoside xylosyltransferase 2 |
| chr7_+_49559859 | 0.13 |
ENSMUST00000056442.12
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr1_-_135032972 | 0.13 |
ENSMUST00000044828.14
|
Lgr6
|
leucine-rich repeat-containing G protein-coupled receptor 6 |
| chr10_-_31485180 | 0.12 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr2_-_33321306 | 0.12 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr12_+_24881582 | 0.12 |
ENSMUST00000221952.2
ENSMUST00000078902.8 ENSMUST00000110942.11 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr10_-_53252210 | 0.11 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr15_-_8473918 | 0.11 |
ENSMUST00000052965.8
|
Nipbl
|
NIPBL cohesin loading factor |
| chr1_-_151965876 | 0.10 |
ENSMUST00000044581.14
|
1700025G04Rik
|
RIKEN cDNA 1700025G04 gene |
| chr2_-_178056251 | 0.10 |
ENSMUST00000058678.5
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr6_+_145156860 | 0.10 |
ENSMUST00000111725.8
ENSMUST00000111726.10 ENSMUST00000039729.5 ENSMUST00000111723.8 ENSMUST00000111724.8 ENSMUST00000111721.2 ENSMUST00000111719.2 |
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chrX_+_23559282 | 0.09 |
ENSMUST00000035766.13
ENSMUST00000101670.3 |
Wdr44
|
WD repeat domain 44 |
| chr9_+_35332837 | 0.09 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr2_+_162773440 | 0.09 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr6_-_120015608 | 0.09 |
ENSMUST00000177761.8
ENSMUST00000088644.13 ENSMUST00000060043.13 |
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr6_-_97594498 | 0.09 |
ENSMUST00000113355.9
|
Frmd4b
|
FERM domain containing 4B |
| chr9_-_44145280 | 0.08 |
ENSMUST00000205968.2
ENSMUST00000206147.2 ENSMUST00000037644.8 |
Cbl
|
Casitas B-lineage lymphoma |
| chr1_+_60219724 | 0.08 |
ENSMUST00000160834.8
|
Nbeal1
|
neurobeachin like 1 |
| chr10_-_10348058 | 0.08 |
ENSMUST00000179956.8
ENSMUST00000208717.2 ENSMUST00000172530.8 ENSMUST00000132573.2 |
Adgb
|
androglobin |
| chr5_+_91222470 | 0.08 |
ENSMUST00000031324.6
|
Ereg
|
epiregulin |
| chr18_-_38068456 | 0.07 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr10_+_19232281 | 0.06 |
ENSMUST00000053225.7
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chr5_+_32616187 | 0.06 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr15_+_6737853 | 0.06 |
ENSMUST00000061656.8
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chrX_-_94488394 | 0.06 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1 |
| chr16_+_35803674 | 0.06 |
ENSMUST00000004054.13
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr10_-_39009844 | 0.05 |
ENSMUST00000134279.8
ENSMUST00000139743.8 ENSMUST00000149949.8 ENSMUST00000124941.8 ENSMUST00000125042.8 ENSMUST00000063204.9 |
Fam229b
|
family with sequence similarity 229, member B |
| chr6_+_65019574 | 0.05 |
ENSMUST00000031984.9
ENSMUST00000205118.3 |
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_-_69034425 | 0.04 |
ENSMUST00000194558.6
ENSMUST00000029353.9 |
Kpna4
|
karyopherin (importin) alpha 4 |
| chr5_+_72805122 | 0.04 |
ENSMUST00000087212.8
|
Nipal1
|
NIPA-like domain containing 1 |
| chr16_+_91855158 | 0.04 |
ENSMUST00000047429.9
ENSMUST00000232677.2 ENSMUST00000113975.3 |
Mrps6
Gm49711
Slc5a3
|
mitochondrial ribosomal protein S6 predicted gene, 49711 solute carrier family 5 (inositol transporters), member 3 |
| chr5_+_107745562 | 0.04 |
ENSMUST00000112651.8
ENSMUST00000112654.8 ENSMUST00000065422.12 |
Rpap2
|
RNA polymerase II associated protein 2 |
| chr1_-_131066004 | 0.04 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr15_-_35155896 | 0.04 |
ENSMUST00000067033.8
ENSMUST00000018476.14 |
Stk3
|
serine/threonine kinase 3 |
| chr4_+_83443777 | 0.03 |
ENSMUST00000053414.13
ENSMUST00000231339.2 ENSMUST00000126429.8 |
Ccdc171
|
coiled-coil domain containing 171 |
| chr9_-_50657800 | 0.02 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr3_+_116306719 | 0.02 |
ENSMUST00000000349.11
ENSMUST00000197201.5 |
Dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr2_+_69652714 | 0.02 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr3_-_41037427 | 0.02 |
ENSMUST00000058578.8
|
Pgrmc2
|
progesterone receptor membrane component 2 |
| chr5_+_88702307 | 0.01 |
ENSMUST00000090413.6
|
Utp3
|
UTP3 small subunit processome component |
| chrX_-_47123719 | 0.01 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr1_-_10302895 | 0.01 |
ENSMUST00000088615.11
ENSMUST00000131556.2 |
Arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) |
| chr14_-_73563212 | 0.01 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.3 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |