Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
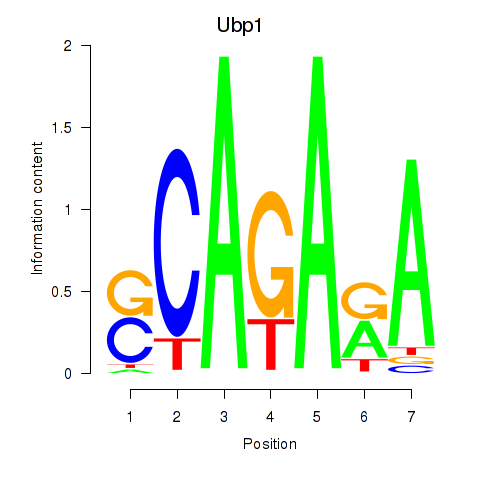
Results for Ubp1
Z-value: 1.46
Transcription factors associated with Ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ubp1
|
ENSMUSG00000009741.16 | Ubp1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ubp1 | mm39_v1_chr9_+_113760002_113760055 | 0.53 | 1.3e-06 | Click! |
Activity profile of Ubp1 motif
Sorted Z-values of Ubp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ubp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_67838100 | 9.95 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr6_+_41523664 | 9.77 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr10_-_111833138 | 8.63 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr7_-_3848050 | 8.54 |
ENSMUST00000108615.10
ENSMUST00000119469.2 |
Pira2
|
paired-Ig-like receptor A2 |
| chr6_-_70149254 | 7.97 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr6_+_70648743 | 7.72 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr6_-_69521891 | 7.68 |
ENSMUST00000103356.4
|
Igkv4-57-1
|
immunoglobulin kappa variable 4-57-1 |
| chr6_+_41107047 | 7.42 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr5_-_113957362 | 7.17 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr6_+_41139948 | 6.82 |
ENSMUST00000103275.4
|
Trbv17
|
T cell receptor beta, variable 17 |
| chr17_+_28426752 | 6.48 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr6_-_70318437 | 6.37 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr6_-_69792108 | 6.29 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr6_-_70383976 | 6.29 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr6_-_70121150 | 6.17 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr12_+_85733415 | 6.13 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr6_-_68713748 | 6.10 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_-_69704122 | 6.01 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr6_-_69753317 | 5.94 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr6_+_41025217 | 5.81 |
ENSMUST00000103264.3
|
Trbv3
|
T cell receptor beta, variable 3 |
| chr19_-_11243530 | 5.74 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_+_70549568 | 5.73 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr6_-_69877642 | 5.70 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr6_-_70051586 | 5.64 |
ENSMUST00000103377.3
|
Igkv6-32
|
immunoglobulin kappa variable 6-32 |
| chr6_+_41515152 | 5.62 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr9_+_51124983 | 5.51 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr16_-_19060440 | 5.40 |
ENSMUST00000103751.3
|
Iglv3
|
immunoglobulin lambda variable 3 |
| chr16_-_18884431 | 5.40 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr16_+_48692976 | 5.36 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr6_-_70021662 | 5.34 |
ENSMUST00000196959.2
|
Igkv8-34
|
immunoglobulin kappa variable 8-34 |
| chr6_+_70495224 | 5.29 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr17_-_57385490 | 5.24 |
ENSMUST00000011623.9
|
Dennd1c
|
DENN/MADD domain containing 1C |
| chr6_+_41118120 | 5.20 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr1_-_192880260 | 5.19 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr13_-_55676334 | 5.14 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr7_-_126817475 | 5.09 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr12_-_113802603 | 4.90 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr17_+_28426831 | 4.84 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr17_+_34524841 | 4.83 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr4_+_130643260 | 4.78 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr1_-_85247864 | 4.77 |
ENSMUST00000159582.8
ENSMUST00000161267.8 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr6_+_70640233 | 4.70 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr6_+_70348416 | 4.70 |
ENSMUST00000103391.4
|
Igkv6-17
|
immunoglobulin kappa variable 6-17 |
| chr6_-_125213911 | 4.69 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr12_+_113115632 | 4.60 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr1_-_138103021 | 4.44 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_70435020 | 4.44 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr2_-_25517945 | 4.34 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr1_+_85528392 | 4.25 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr11_-_16958647 | 4.22 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr6_-_70217956 | 4.17 |
ENSMUST00000196275.2
|
Gm43218
|
predicted gene 43218 |
| chr6_-_124865155 | 4.16 |
ENSMUST00000024044.7
|
Cd4
|
CD4 antigen |
| chr6_-_69877961 | 4.14 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr5_-_113957318 | 4.11 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr9_+_32547529 | 4.10 |
ENSMUST00000238819.2
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr15_+_6673167 | 4.09 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr12_-_114901026 | 4.06 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr10_-_81335966 | 4.05 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr6_-_68907718 | 4.05 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr17_+_48571298 | 4.04 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr9_+_43956969 | 4.04 |
ENSMUST00000215809.2
|
Thy1
|
thymus cell antigen 1, theta |
| chr6_-_129252396 | 3.98 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr6_-_69415741 | 3.96 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr10_-_21036824 | 3.96 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr6_-_130208601 | 3.96 |
ENSMUST00000088011.11
ENSMUST00000112013.8 ENSMUST00000049304.14 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr6_+_72281587 | 3.93 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr1_-_133617824 | 3.92 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr7_-_126303887 | 3.83 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_+_142030744 | 3.81 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr17_+_34524884 | 3.79 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr11_-_79421397 | 3.78 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chrX_+_149330371 | 3.76 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr2_-_118558825 | 3.74 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr9_-_32454157 | 3.73 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr14_+_53921052 | 3.69 |
ENSMUST00000103663.6
|
Trav4-4-dv10
|
T cell receptor alpha variable 4-4-DV10 |
| chr10_-_117128763 | 3.67 |
ENSMUST00000092162.7
|
Lyz1
|
lysozyme 1 |
| chr6_+_68547717 | 3.61 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr6_+_41155309 | 3.60 |
ENSMUST00000103276.3
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr7_+_4340708 | 3.59 |
ENSMUST00000006792.6
ENSMUST00000126417.3 |
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr1_-_170755109 | 3.57 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
| chr6_+_41039255 | 3.56 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr6_+_41519654 | 3.50 |
ENSMUST00000103293.2
|
Trbj2-1
|
T cell receptor beta joining 2-1 |
| chr5_+_90920294 | 3.49 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr2_+_90912710 | 3.45 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr12_-_114012399 | 3.44 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr6_+_38895902 | 3.42 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr9_+_110867807 | 3.41 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr11_+_95728042 | 3.41 |
ENSMUST00000107712.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr4_+_114914880 | 3.41 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr9_+_43957241 | 3.39 |
ENSMUST00000214627.2
|
Thy1
|
thymus cell antigen 1, theta |
| chr11_+_33997114 | 3.36 |
ENSMUST00000109329.9
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr2_-_164198427 | 3.36 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr11_+_72851989 | 3.34 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr12_-_113680723 | 3.32 |
ENSMUST00000179657.2
|
Ighv2-6
|
immunoglobulin heavy variable 2-6 |
| chr7_+_55417967 | 3.32 |
ENSMUST00000060416.15
ENSMUST00000094360.13 ENSMUST00000165045.9 ENSMUST00000173835.2 |
Siglech
|
sialic acid binding Ig-like lectin H |
| chr12_-_114023935 | 3.29 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr6_-_69204417 | 3.29 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr17_-_50401305 | 3.24 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr16_-_45940602 | 3.24 |
ENSMUST00000023336.10
|
Cd96
|
CD96 antigen |
| chrX_-_133062677 | 3.22 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr15_+_78128990 | 3.20 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr11_-_52173391 | 3.20 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr17_+_34544632 | 3.18 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr5_+_90920353 | 3.18 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr6_+_41112064 | 3.16 |
ENSMUST00000103272.4
|
Trbv14
|
T cell receptor beta, variable 14 |
| chr5_-_134258435 | 3.14 |
ENSMUST00000016094.13
ENSMUST00000111275.8 ENSMUST00000144086.2 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr17_+_34416707 | 3.13 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr2_-_118558852 | 3.10 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr9_+_20927271 | 3.09 |
ENSMUST00000086399.6
|
Icam1
|
intercellular adhesion molecule 1 |
| chr16_-_75706161 | 3.09 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr8_-_79539838 | 3.09 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr13_+_19369097 | 3.07 |
ENSMUST00000103554.5
|
Trgv4
|
T cell receptor gamma, variable 4 |
| chr12_-_113589576 | 3.05 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr7_-_43309563 | 3.05 |
ENSMUST00000032667.10
|
Siglece
|
sialic acid binding Ig-like lectin E |
| chr6_-_129252323 | 3.04 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr6_-_70120881 | 3.03 |
ENSMUST00000103380.3
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chrX_-_100312629 | 2.99 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr6_-_70364222 | 2.99 |
ENSMUST00000103392.3
ENSMUST00000195945.2 |
Igkv8-16
|
immunoglobulin kappa variable 8-16 |
| chr10_+_51367052 | 2.98 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chr6_-_68746087 | 2.98 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr6_-_68784692 | 2.97 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr17_+_32755525 | 2.92 |
ENSMUST00000169591.8
ENSMUST00000003416.15 |
Cyp4f16
|
cytochrome P450, family 4, subfamily f, polypeptide 16 |
| chr6_-_70313491 | 2.91 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr7_+_28682253 | 2.91 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr11_+_104468107 | 2.89 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr15_+_4756684 | 2.87 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr19_+_45064539 | 2.86 |
ENSMUST00000026234.5
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr2_-_129213050 | 2.81 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr10_+_39009951 | 2.81 |
ENSMUST00000019991.8
|
Tube1
|
tubulin, epsilon 1 |
| chr6_-_68887922 | 2.80 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr12_-_114621406 | 2.79 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr8_+_57964921 | 2.78 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr7_+_67925718 | 2.78 |
ENSMUST00000210558.2
|
Fam169b
|
family with sequence similarity 169, member B |
| chr12_+_76371634 | 2.75 |
ENSMUST00000154078.3
ENSMUST00000095610.9 |
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr12_-_114117264 | 2.75 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr13_-_12535236 | 2.73 |
ENSMUST00000179308.3
|
Edaradd
|
EDAR (ectodysplasin-A receptor)-associated death domain |
| chrX_-_47551990 | 2.73 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr12_-_115157739 | 2.72 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr11_-_70703365 | 2.70 |
ENSMUST00000074572.7
ENSMUST00000108534.9 |
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr8_+_27468175 | 2.67 |
ENSMUST00000209411.2
|
Zfp703
|
zinc finger protein 703 |
| chr17_+_37180437 | 2.66 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr14_-_56339915 | 2.64 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chrX_+_158491589 | 2.62 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr4_-_149747644 | 2.61 |
ENSMUST00000105689.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr12_-_33142858 | 2.60 |
ENSMUST00000095774.3
|
Cdhr3
|
cadherin-related family member 3 |
| chr3_+_134918298 | 2.60 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr1_+_60948149 | 2.59 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr15_+_78129040 | 2.59 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr12_-_114286421 | 2.58 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr7_+_78563150 | 2.56 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr11_-_103158190 | 2.55 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr11_+_45946800 | 2.52 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr7_+_87927293 | 2.52 |
ENSMUST00000032779.12
ENSMUST00000131108.9 ENSMUST00000128791.2 |
Ctsc
|
cathepsin C |
| chr9_-_37166699 | 2.52 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr15_-_100583044 | 2.51 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr16_+_48662894 | 2.51 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr12_-_113733922 | 2.51 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr2_+_120838884 | 2.51 |
ENSMUST00000060455.15
ENSMUST00000099488.11 ENSMUST00000099489.9 ENSMUST00000171260.3 |
Ccndbp1
|
cyclin D-type binding-protein 1 |
| chrX_-_106446928 | 2.51 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chrX_-_166047275 | 2.50 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr2_+_103799873 | 2.50 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr7_-_140596811 | 2.50 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr6_+_38918327 | 2.49 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr6_-_83010402 | 2.46 |
ENSMUST00000089651.6
|
Dok1
|
docking protein 1 |
| chr4_-_156340276 | 2.46 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr8_+_57964956 | 2.45 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr7_-_3901119 | 2.43 |
ENSMUST00000070639.8
|
Gm14548
|
predicted gene 14548 |
| chr1_-_131161312 | 2.42 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr17_+_21910767 | 2.40 |
ENSMUST00000072133.5
|
Gm10226
|
predicted gene 10226 |
| chr2_+_84629172 | 2.39 |
ENSMUST00000102642.9
ENSMUST00000150325.2 |
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr15_+_44320918 | 2.39 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr14_+_53903370 | 2.38 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr12_-_115031622 | 2.38 |
ENSMUST00000194257.2
|
Ighv8-5
|
immunoglobulin heavy variable V8-5 |
| chr5_-_72325482 | 2.37 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chr1_-_85248264 | 2.37 |
ENSMUST00000093506.12
ENSMUST00000064341.9 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr14_+_14475188 | 2.36 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr12_-_113666198 | 2.35 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr11_+_117740111 | 2.35 |
ENSMUST00000093906.5
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr6_-_70036183 | 2.33 |
ENSMUST00000197429.5
ENSMUST00000103376.3 |
Igkv7-33
|
immunoglobulin kappa chain variable 7-33 |
| chr6_-_68840015 | 2.33 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr2_+_174602412 | 2.32 |
ENSMUST00000029030.9
|
Edn3
|
endothelin 3 |
| chr7_-_128020397 | 2.32 |
ENSMUST00000147840.2
|
Rgs10
|
regulator of G-protein signalling 10 |
| chr3_-_19319155 | 2.31 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr14_+_60970355 | 2.31 |
ENSMUST00000162945.2
|
Spata13
|
spermatogenesis associated 13 |
| chr6_+_70332836 | 2.29 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr10_+_19497740 | 2.28 |
ENSMUST00000036564.8
|
Il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr1_-_143879877 | 2.26 |
ENSMUST00000127206.8
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr10_-_14420725 | 2.26 |
ENSMUST00000041168.6
ENSMUST00000238680.2 |
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr2_-_31973795 | 2.26 |
ENSMUST00000056406.7
|
Fam78a
|
family with sequence similarity 78, member A |
| chr13_+_19524136 | 2.25 |
ENSMUST00000103564.3
|
Trgv1
|
T cell receptor gamma, variable 1 |
| chr17_-_36440317 | 2.25 |
ENSMUST00000046131.16
ENSMUST00000173322.8 ENSMUST00000172968.2 |
Gm7030
|
predicted gene 7030 |
| chr19_+_29229826 | 2.24 |
ENSMUST00000238009.2
|
Jak2
|
Janus kinase 2 |
| chr16_-_20245071 | 2.22 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_-_7086018 | 2.21 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_+_140427799 | 2.18 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr7_+_78563513 | 2.18 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chrX_+_8137881 | 2.18 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr6_+_30639217 | 2.17 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr12_-_113928438 | 2.16 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr7_-_44785480 | 2.16 |
ENSMUST00000211246.2
ENSMUST00000210197.2 |
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 2.0 | 10.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.4 | 4.2 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 1.3 | 10.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 1.2 | 3.7 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.1 | 3.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.1 | 2.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 1.1 | 4.4 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 1.0 | 3.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.9 | 6.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.9 | 3.4 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.8 | 4.9 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.8 | 4.8 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.8 | 4.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 0.8 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.8 | 3.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.8 | 5.3 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.7 | 2.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.7 | 2.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.7 | 3.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.7 | 2.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.7 | 2.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.7 | 4.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.7 | 2.7 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.7 | 4.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.6 | 3.2 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.6 | 3.8 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.6 | 4.4 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.6 | 3.7 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.6 | 10.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.6 | 155.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.6 | 3.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 7.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.5 | 1.6 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.5 | 1.6 | GO:0071724 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.5 | 2.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.5 | 5.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.5 | 5.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 2.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.5 | 1.4 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.5 | 2.9 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.5 | 3.8 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.5 | 68.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 1.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 1.3 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.4 | 2.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.4 | 0.9 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.4 | 3.0 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.4 | 1.3 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.4 | 2.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.4 | 1.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 2.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.4 | 1.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.4 | 2.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 | 1.5 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.4 | 2.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 6.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 0.7 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.4 | 1.4 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.3 | 2.4 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.3 | 4.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 2.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 1.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.3 | 2.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.3 | 2.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.3 | 2.6 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.3 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 1.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.3 | 1.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.3 | 3.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 1.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.3 | 1.0 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.3 | 1.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.3 | 0.9 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.3 | 1.2 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.3 | 1.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 4.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 2.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 1.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.3 | 1.9 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.3 | 7.4 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.3 | 1.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 0.8 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.3 | 0.8 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.3 | 1.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.3 | 1.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.3 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.3 | 1.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 1.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.3 | 1.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.3 | 1.5 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 2.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 1.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 2.6 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 2.8 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 4.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 1.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 3.6 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.2 | 2.7 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.2 | 0.9 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 1.1 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.2 | 7.4 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.2 | 1.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.2 | 2.5 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 0.8 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 0.9 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.2 | 0.7 | GO:0035549 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.2 | 1.3 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 0.7 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 1.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 3.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.2 | 2.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 2.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 0.9 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.2 | 1.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.5 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.2 | 1.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 2.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 1.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.1 | 0.9 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 2.8 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 5.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 2.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.7 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 2.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 0.5 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.9 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 1.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 3.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.1 | 0.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.4 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 2.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 2.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 3.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 2.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.5 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 2.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 4.5 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 1.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.9 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.9 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 1.4 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 3.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.1 | 1.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.2 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 2.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.4 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 4.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.1 | 0.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.6 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.3 | GO:0061643 | vagus nerve morphogenesis(GO:0021644) chemorepulsion of axon(GO:0061643) |
| 0.1 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.3 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.0 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 1.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 6.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 2.0 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.8 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 2.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.6 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 5.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.7 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 3.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 5.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 1.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.2 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 4.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 0.4 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.5 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 1.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 1.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.5 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 1.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.0 | 2.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.3 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0070814 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 2.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 1.7 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) |
| 0.0 | 1.0 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 1.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 1.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.1 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 2.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.2 | GO:0048524 | positive regulation of viral process(GO:0048524) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 1.6 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.0 | 0.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 3.3 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.4 | GO:0042107 | cytokine metabolic process(GO:0042107) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.6 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.0 | 1.4 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.0 | 0.8 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 1.7 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 1.9 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.1 | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043122) |
| 0.0 | 0.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.1 | 3.4 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.1 | 3.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.8 | 14.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.7 | 8.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 4.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 2.3 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.6 | 1.7 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.6 | 3.4 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.6 | 2.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.6 | 2.8 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.6 | 68.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 3.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 6.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.5 | 1.9 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.4 | 1.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.6 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.4 | 4.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 10.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 2.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 3.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 2.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 1.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 2.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 3.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 4.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 1.1 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.3 | 1.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.9 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 1.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 0.7 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.2 | 2.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 4.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 2.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 1.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 0.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.6 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 2.0 | GO:1990712 | extrinsic component of external side of plasma membrane(GO:0031232) HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 9.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 7.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.6 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 2.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 9.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 44.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.8 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 1.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.2 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 3.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.9 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.3 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 1.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 2.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 104.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 5.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 8.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 4.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 2.0 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 2.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 6.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.9 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 3.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 7.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 3.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 8.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.1 | 6.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 4.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.0 | 2.9 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 4.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 2.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.8 | 4.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.8 | 6.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.8 | 9.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 4.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 2.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.7 | 4.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 1.9 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.6 | 2.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.6 | 6.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 1.6 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.5 | 1.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.5 | 5.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 68.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.5 | 1.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 2.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 1.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.4 | 1.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.4 | 2.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.4 | 1.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.4 | 0.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 2.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 1.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.4 | 1.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 1.5 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.4 | 2.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 2.5 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 1.7 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 1.0 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.3 | 1.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 1.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 5.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 2.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 4.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 2.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 1.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 2.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 1.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 1.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 1.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.3 | 10.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 4.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 4.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 6.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 1.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 0.8 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 0.7 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.2 | 0.7 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.2 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 5.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.2 | 0.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 5.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 3.5 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.2 | 0.8 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 2.6 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 3.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.0 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 2.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 2.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 0.7 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 4.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 0.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 0.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 4.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 11.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 1.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 1.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.9 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 1.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 2.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.4 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 7.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 3.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 11.9 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.0 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 7.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 5.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.3 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 4.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 2.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 2.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 7.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.5 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 3.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.4 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 2.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 1.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 6.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 3.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 2.1 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 3.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 3.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 1.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.0 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 3.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 3.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0030881 | IgG receptor activity(GO:0019770) beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 7.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 3.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 1.4 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 0.4 | GO:0005402 | cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 3.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 8.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 11.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 3.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 20.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 2.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 19.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 4.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 11.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 9.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 2.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 11.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 5.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 4.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 3.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 5.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 3.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 6.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 6.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 4.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 4.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 5.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 4.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 7.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 4.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 1.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 3.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 5.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 4.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 3.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 3.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 1.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 2.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.9 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 1.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 8.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 6.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 2.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 8.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 5.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.9 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 13.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 4.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 10.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 6.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 7.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 7.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 3.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |