Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Vdr
Z-value: 1.08
Transcription factors associated with Vdr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vdr
|
ENSMUSG00000022479.16 | Vdr |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vdr | mm39_v1_chr15_-_97808502_97808523 | -0.25 | 3.2e-02 | Click! |
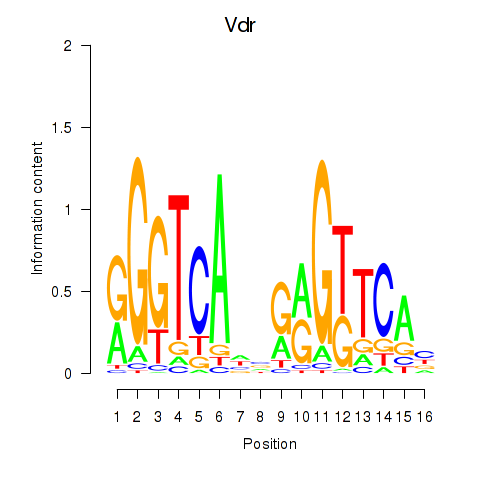
Activity profile of Vdr motif
Sorted Z-values of Vdr motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Vdr
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_123077515 | 7.96 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr10_+_126914755 | 7.70 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr15_+_82159094 | 7.65 |
ENSMUST00000116423.3
ENSMUST00000230418.2 |
Septin3
|
septin 3 |
| chr16_-_94657531 | 7.24 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr4_-_129015493 | 6.60 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr11_-_116303791 | 5.80 |
ENSMUST00000100202.10
ENSMUST00000106398.9 |
Rnf157
|
ring finger protein 157 |
| chr7_-_139734637 | 5.47 |
ENSMUST00000059241.8
|
Sprn
|
shadow of prion protein |
| chr15_+_82159398 | 5.23 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chrX_+_135491587 | 5.20 |
ENSMUST00000138047.8
ENSMUST00000145648.8 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr4_+_123077286 | 5.20 |
ENSMUST00000126995.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr18_+_82493284 | 5.17 |
ENSMUST00000047865.14
|
Mbp
|
myelin basic protein |
| chr8_+_96429665 | 5.16 |
ENSMUST00000073139.14
ENSMUST00000080666.8 ENSMUST00000212160.2 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr11_-_3672188 | 4.96 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr2_+_148237258 | 4.95 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr9_-_37464200 | 4.95 |
ENSMUST00000065668.12
|
Nrgn
|
neurogranin |
| chr1_-_180310894 | 4.87 |
ENSMUST00000211561.2
ENSMUST00000136521.2 ENSMUST00000179826.2 |
Stum
|
mechanosensory transduction mediator |
| chr5_-_71815318 | 4.58 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr6_+_125192514 | 4.56 |
ENSMUST00000032487.14
ENSMUST00000100942.9 ENSMUST00000063588.11 |
Vamp1
|
vesicle-associated membrane protein 1 |
| chr7_-_27374017 | 4.42 |
ENSMUST00000036453.14
ENSMUST00000108341.2 |
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr5_-_8417982 | 4.38 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr1_-_5089564 | 4.35 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr13_-_12121831 | 4.34 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr8_+_70782467 | 4.32 |
ENSMUST00000207684.2
|
Gdf1
|
growth differentiation factor 1 |
| chr11_-_103829040 | 4.24 |
ENSMUST00000133774.4
ENSMUST00000149642.3 |
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr1_+_66403774 | 4.16 |
ENSMUST00000077355.12
ENSMUST00000114012.8 |
Map2
|
microtubule-associated protein 2 |
| chr4_+_148245070 | 4.02 |
ENSMUST00000047951.9
|
Fbxo2
|
F-box protein 2 |
| chr14_+_9646630 | 3.93 |
ENSMUST00000112658.8
ENSMUST00000112657.9 ENSMUST00000177814.2 ENSMUST00000067491.14 |
Cadps
|
Ca2+-dependent secretion activator |
| chr1_+_159351337 | 3.71 |
ENSMUST00000192069.6
|
Tnr
|
tenascin R |
| chr11_-_83957889 | 3.70 |
ENSMUST00000108101.8
|
Dusp14
|
dual specificity phosphatase 14 |
| chr11_+_72909811 | 3.61 |
ENSMUST00000092937.13
|
Camkk1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr7_-_25374472 | 3.56 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr13_-_95581335 | 3.53 |
ENSMUST00000045583.9
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr13_-_95581393 | 3.49 |
ENSMUST00000221025.2
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr8_+_36232861 | 3.43 |
ENSMUST00000207505.2
|
Gm19410
|
predicted gene, 19410 |
| chr1_+_135710803 | 3.42 |
ENSMUST00000132795.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr13_+_105580147 | 3.39 |
ENSMUST00000022235.6
|
Htr1a
|
5-hydroxytryptamine (serotonin) receptor 1A |
| chr7_-_44861235 | 3.36 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr9_-_86762467 | 3.36 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr10_+_127561259 | 3.27 |
ENSMUST00000026466.5
|
Tac2
|
tachykinin 2 |
| chr18_+_33072194 | 3.23 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr7_+_43474819 | 3.14 |
ENSMUST00000107967.3
|
Klk6
|
kallikrein related-peptidase 6 |
| chr10_+_57661010 | 3.11 |
ENSMUST00000165013.2
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr7_-_141927471 | 3.07 |
ENSMUST00000105988.2
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr10_+_57660948 | 3.05 |
ENSMUST00000020024.12
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr9_+_87026337 | 3.02 |
ENSMUST00000113149.8
ENSMUST00000049457.14 ENSMUST00000179313.3 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr8_-_106015682 | 2.98 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr15_+_82140224 | 2.97 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr11_+_3438274 | 2.92 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr17_-_43003135 | 2.90 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr1_-_135934080 | 2.85 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr8_-_106016097 | 2.76 |
ENSMUST00000171788.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr14_-_51134930 | 2.72 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr11_+_54195006 | 2.71 |
ENSMUST00000108904.10
ENSMUST00000108905.10 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_-_180681079 | 2.70 |
ENSMUST00000067120.14
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr9_+_110595224 | 2.65 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr3_+_95067759 | 2.62 |
ENSMUST00000131742.8
ENSMUST00000090823.8 ENSMUST00000090821.10 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr6_+_4902912 | 2.61 |
ENSMUST00000175889.8
ENSMUST00000168998.9 |
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr4_-_43578823 | 2.60 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr1_+_92834681 | 2.58 |
ENSMUST00000060913.8
|
Dusp28
|
dual specificity phosphatase 28 |
| chr17_+_46564457 | 2.55 |
ENSMUST00000233692.2
|
Lrrc73
|
leucine rich repeat containing 73 |
| chrX_+_7744535 | 2.54 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr6_-_113478779 | 2.49 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr4_+_156300325 | 2.42 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr11_+_58854616 | 2.41 |
ENSMUST00000075141.7
|
Trim17
|
tripartite motif-containing 17 |
| chr7_-_16651107 | 2.40 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr7_+_126376353 | 2.39 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr7_+_27259895 | 2.38 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr7_+_126376099 | 2.37 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr7_-_27373939 | 2.34 |
ENSMUST00000138243.2
|
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr19_+_47167259 | 2.33 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr4_-_43578636 | 2.31 |
ENSMUST00000130443.2
|
Gba2
|
glucosidase beta 2 |
| chr16_-_18052937 | 2.21 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr5_-_124233812 | 2.18 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr2_-_180680868 | 2.14 |
ENSMUST00000108851.8
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr15_-_94302139 | 2.13 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr3_+_87855973 | 2.13 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr7_+_4463686 | 2.11 |
ENSMUST00000167298.2
ENSMUST00000171445.8 |
Eps8l1
|
EPS8-like 1 |
| chr15_-_97629209 | 2.10 |
ENSMUST00000100249.10
|
Endou
|
endonuclease, polyU-specific |
| chr4_-_140393185 | 2.08 |
ENSMUST00000069623.12
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr6_+_86605146 | 2.04 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr7_+_43473943 | 2.01 |
ENSMUST00000107966.10
ENSMUST00000177514.8 |
Klk6
|
kallikrein related-peptidase 6 |
| chr14_-_76794329 | 1.93 |
ENSMUST00000228524.2
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr7_+_127347339 | 1.92 |
ENSMUST00000206893.2
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr7_-_19043955 | 1.91 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chr6_+_134617903 | 1.91 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr3_-_91990439 | 1.89 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chr5_+_110248276 | 1.88 |
ENSMUST00000141066.8
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr18_+_57266123 | 1.88 |
ENSMUST00000075770.13
|
Megf10
|
multiple EGF-like-domains 10 |
| chr16_-_18448454 | 1.88 |
ENSMUST00000231622.2
|
Septin5
|
septin 5 |
| chr10_+_79552421 | 1.85 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr12_-_84408576 | 1.85 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr5_-_136911969 | 1.84 |
ENSMUST00000057497.13
ENSMUST00000111103.2 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr7_+_29008840 | 1.84 |
ENSMUST00000137848.2
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr3_-_108133914 | 1.79 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr5_-_23881353 | 1.77 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chrX_-_48877080 | 1.76 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr9_-_86762450 | 1.76 |
ENSMUST00000191290.3
|
Snap91
|
synaptosomal-associated protein 91 |
| chr7_-_140480314 | 1.73 |
ENSMUST00000026561.10
|
Cox8b
|
cytochrome c oxidase subunit 8B |
| chr5_-_121641461 | 1.71 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr12_-_111638722 | 1.68 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr6_+_4903299 | 1.67 |
ENSMUST00000035813.9
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr1_+_75498162 | 1.57 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr2_-_111779785 | 1.57 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr9_+_117869543 | 1.55 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr17_-_33932346 | 1.53 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr5_-_115236354 | 1.52 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr7_+_43473917 | 1.52 |
ENSMUST00000107968.10
|
Klk6
|
kallikrein related-peptidase 6 |
| chr11_+_83189831 | 1.51 |
ENSMUST00000176944.8
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr14_-_51134906 | 1.51 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr2_+_32498716 | 1.50 |
ENSMUST00000129165.2
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr14_-_70666513 | 1.50 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr7_-_12156678 | 1.50 |
ENSMUST00000080348.12
|
Zfp551
|
zinc fingr protein 551 |
| chr10_+_40505985 | 1.50 |
ENSMUST00000019977.8
ENSMUST00000214102.2 ENSMUST00000213503.2 ENSMUST00000213442.2 ENSMUST00000216830.2 |
Ddo
|
D-aspartate oxidase |
| chr5_+_101912939 | 1.47 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr9_+_53678801 | 1.43 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr7_+_127347308 | 1.43 |
ENSMUST00000188580.3
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr12_-_87312994 | 1.39 |
ENSMUST00000072744.15
|
Vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr18_-_13074834 | 1.38 |
ENSMUST00000122175.8
ENSMUST00000142467.2 ENSMUST00000074352.11 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr9_-_21159733 | 1.37 |
ENSMUST00000122088.2
|
S1pr5
|
sphingosine-1-phosphate receptor 5 |
| chr8_-_123187406 | 1.37 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr2_-_25124240 | 1.37 |
ENSMUST00000006638.8
|
Slc34a3
|
solute carrier family 34 (sodium phosphate), member 3 |
| chr19_-_8775935 | 1.34 |
ENSMUST00000096261.5
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr9_+_110592709 | 1.33 |
ENSMUST00000079784.12
|
Myl3
|
myosin, light polypeptide 3 |
| chr4_-_42874178 | 1.33 |
ENSMUST00000107978.2
|
Fam205c
|
family with sequence similarity 205, member C |
| chr5_+_136721938 | 1.33 |
ENSMUST00000196068.5
ENSMUST00000005611.10 |
Myl10
|
myosin, light chain 10, regulatory |
| chrX_-_58612709 | 1.32 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_+_136013372 | 1.32 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr9_+_57411279 | 1.31 |
ENSMUST00000080514.9
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr8_+_124170027 | 1.30 |
ENSMUST00000108830.2
|
Def8
|
differentially expressed in FDCP 8 |
| chr17_-_25155868 | 1.30 |
ENSMUST00000115228.9
ENSMUST00000117509.8 ENSMUST00000121723.8 ENSMUST00000119115.8 ENSMUST00000121787.8 ENSMUST00000088345.12 ENSMUST00000120035.8 ENSMUST00000115229.10 ENSMUST00000178969.8 |
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr4_-_42874197 | 1.27 |
ENSMUST00000055944.11
|
Fam205c
|
family with sequence similarity 205, member C |
| chr9_-_107544573 | 1.26 |
ENSMUST00000010208.14
ENSMUST00000193932.6 |
Slc38a3
|
solute carrier family 38, member 3 |
| chr17_+_48006096 | 1.26 |
ENSMUST00000113296.8
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr9_+_121126562 | 1.26 |
ENSMUST00000238988.2
ENSMUST00000210798.3 ENSMUST00000211439.2 |
Trak1
|
trafficking protein, kinesin binding 1 |
| chr5_+_36050663 | 1.25 |
ENSMUST00000064571.11
|
Afap1
|
actin filament associated protein 1 |
| chr9_+_117869577 | 1.25 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr12_+_82216800 | 1.23 |
ENSMUST00000222298.2
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr12_-_87312982 | 1.23 |
ENSMUST00000179379.9
|
Vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr16_+_17884267 | 1.23 |
ENSMUST00000151266.8
ENSMUST00000066027.14 ENSMUST00000155387.8 |
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr16_-_31094095 | 1.22 |
ENSMUST00000060188.14
|
Ppp1r2
|
protein phosphatase 1, regulatory inhibitor subunit 2 |
| chr17_+_34148485 | 1.21 |
ENSMUST00000047503.16
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr18_+_82493237 | 1.20 |
ENSMUST00000091789.11
ENSMUST00000114676.8 |
Mbp
|
myelin basic protein |
| chr17_-_24369884 | 1.20 |
ENSMUST00000052462.13
|
Pdpk1
|
3-phosphoinositide dependent protein kinase 1 |
| chr18_-_7004717 | 1.19 |
ENSMUST00000079788.7
|
Mkx
|
mohawk homeobox |
| chr2_-_165996716 | 1.16 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr10_+_81469522 | 1.15 |
ENSMUST00000140345.2
ENSMUST00000126323.2 |
Ankrd24
|
ankyrin repeat domain 24 |
| chr19_+_47167554 | 1.15 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr11_-_120520954 | 1.14 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr17_+_47815998 | 1.13 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr19_-_8775817 | 1.12 |
ENSMUST00000235964.2
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr8_+_75760301 | 1.11 |
ENSMUST00000165630.3
ENSMUST00000212651.2 ENSMUST00000212388.2 ENSMUST00000212299.2 ENSMUST00000078847.13 ENSMUST00000211869.2 |
Tom1
|
target of myb1 trafficking protein |
| chr7_-_12156516 | 1.11 |
ENSMUST00000120220.3
|
Zfp551
|
zinc fingr protein 551 |
| chr8_+_124169707 | 1.11 |
ENSMUST00000093049.10
ENSMUST00000065534.10 ENSMUST00000001522.10 ENSMUST00000124741.8 ENSMUST00000108832.8 ENSMUST00000132063.8 ENSMUST00000128424.8 |
Def8
|
differentially expressed in FDCP 8 |
| chr6_+_83720022 | 1.11 |
ENSMUST00000206911.2
ENSMUST00000205763.2 |
Atp6v1b1
|
ATPase, H+ transporting, lysosomal V1 subunit B1 |
| chr2_-_174305856 | 1.10 |
ENSMUST00000016396.8
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr15_-_79326200 | 1.08 |
ENSMUST00000117786.9
ENSMUST00000120859.9 |
Csnk1e
|
casein kinase 1, epsilon |
| chr2_-_25507680 | 1.08 |
ENSMUST00000028309.4
|
Ccdc183
|
coiled-coil domain containing 183 |
| chr9_-_72399221 | 1.08 |
ENSMUST00000185151.8
ENSMUST00000085358.12 ENSMUST00000184125.8 ENSMUST00000183574.8 ENSMUST00000184831.8 |
Tex9
|
testis expressed gene 9 |
| chr7_+_5083212 | 1.05 |
ENSMUST00000098845.10
ENSMUST00000146317.8 ENSMUST00000153169.2 ENSMUST00000045277.7 |
Epn1
|
epsin 1 |
| chr11_+_58488171 | 1.05 |
ENSMUST00000054683.8
|
Olfr324
|
olfactory receptor 324 |
| chr3_-_98417830 | 1.04 |
ENSMUST00000196861.2
|
Hsd3b4
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 4 |
| chr10_-_18421796 | 1.03 |
ENSMUST00000214548.2
ENSMUST00000020000.7 |
Hebp2
|
heme binding protein 2 |
| chr17_-_8046125 | 1.02 |
ENSMUST00000239425.2
ENSMUST00000167580.8 |
Fndc1
|
fibronectin type III domain containing 1 |
| chr7_+_6463510 | 1.01 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr5_+_122988111 | 0.99 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chr12_-_103208402 | 0.99 |
ENSMUST00000074416.10
|
Prima1
|
proline rich membrane anchor 1 |
| chr10_-_125164826 | 0.99 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr5_-_129916283 | 0.97 |
ENSMUST00000094280.4
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr11_-_61065846 | 0.96 |
ENSMUST00000041683.9
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr8_+_4303067 | 0.96 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr9_+_53757448 | 0.95 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr2_-_30305401 | 0.94 |
ENSMUST00000142096.2
|
Crat
|
carnitine acetyltransferase |
| chr7_+_15832383 | 0.94 |
ENSMUST00000006181.7
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chr3_+_40801405 | 0.94 |
ENSMUST00000108078.9
|
Abhd18
|
abhydrolase domain containing 18 |
| chr5_-_31359276 | 0.94 |
ENSMUST00000031562.11
|
Zfp513
|
zinc finger protein 513 |
| chr10_-_78300802 | 0.91 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr17_+_34148868 | 0.90 |
ENSMUST00000173266.8
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr8_+_106607507 | 0.85 |
ENSMUST00000040254.16
ENSMUST00000119261.8 |
Edc4
|
enhancer of mRNA decapping 4 |
| chr1_+_172784335 | 0.83 |
ENSMUST00000038432.7
|
Olfr16
|
olfactory receptor 16 |
| chrX_-_103457431 | 0.82 |
ENSMUST00000033695.6
|
Abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr11_-_61652866 | 0.81 |
ENSMUST00000004955.14
ENSMUST00000168115.8 |
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr18_+_38552011 | 0.81 |
ENSMUST00000025293.5
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr14_-_45715308 | 0.79 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr2_+_31840340 | 0.78 |
ENSMUST00000148056.4
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr4_+_102848981 | 0.78 |
ENSMUST00000140654.9
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr4_-_115638209 | 0.77 |
ENSMUST00000106521.2
|
Tex38
|
testis expressed 38 |
| chr10_-_18421628 | 0.76 |
ENSMUST00000213153.2
|
Hebp2
|
heme binding protein 2 |
| chr19_-_58443012 | 0.75 |
ENSMUST00000129100.8
ENSMUST00000123957.2 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr17_-_45744637 | 0.74 |
ENSMUST00000024727.10
|
Cdc5l
|
cell division cycle 5-like (S. pombe) |
| chr17_+_57024759 | 0.73 |
ENSMUST00000002452.8
ENSMUST00000233832.2 ENSMUST00000233233.2 |
Ndufa11
|
NADH:ubiquinone oxidoreductase subunit A11 |
| chr19_+_47167444 | 0.73 |
ENSMUST00000235326.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr18_+_38551960 | 0.73 |
ENSMUST00000236085.2
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr5_-_148336711 | 0.72 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_-_30305313 | 0.72 |
ENSMUST00000132981.9
ENSMUST00000129494.2 |
Crat
|
carnitine acetyltransferase |
| chr7_+_3620356 | 0.70 |
ENSMUST00000076657.11
ENSMUST00000108644.8 |
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr4_-_118037143 | 0.70 |
ENSMUST00000050288.9
ENSMUST00000106403.8 |
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr7_+_107166653 | 0.69 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr17_+_47815968 | 0.68 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr10_-_7668560 | 0.68 |
ENSMUST00000065124.2
|
Ginm1
|
glycoprotein integral membrane 1 |
| chr17_+_28396370 | 0.67 |
ENSMUST00000002318.8
ENSMUST00000233904.2 |
Zfp523
|
zinc finger protein 523 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.8 | 7.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 1.6 | 4.8 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 1.6 | 6.4 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.4 | 4.3 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 1.4 | 4.3 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.0 | 4.9 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.9 | 5.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.8 | 3.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.7 | 4.6 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.7 | 3.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 3.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.6 | 2.8 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 2.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.5 | 1.5 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.5 | 4.6 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.4 | 1.3 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 0.4 | 3.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.4 | 7.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 | 1.5 | GO:0000239 | pachytene(GO:0000239) |
| 0.4 | 5.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 6.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 0.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.3 | 0.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 1.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 1.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.3 | 1.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 1.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.3 | 2.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 1.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.5 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 | 1.8 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.3 | 1.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.3 | 1.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 7.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.2 | 4.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 1.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 6.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 0.7 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 2.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 0.6 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.2 | 2.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.2 | 1.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.6 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.2 | 1.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.5 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 1.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 3.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 4.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 6.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 4.9 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 3.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.0 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 | 2.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 2.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 6.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 4.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.9 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 2.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 7.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 2.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.9 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 4.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 3.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 4.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 2.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 4.6 | GO:0030800 | negative regulation of cyclic nucleotide metabolic process(GO:0030800) |
| 0.1 | 0.4 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 1.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 4.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 1.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 3.0 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 1.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 2.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.0 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.5 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 2.5 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 1.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 1.0 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 1.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 2.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.7 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.7 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.2 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 2.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 1.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 3.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.9 | 3.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.9 | 7.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.9 | 4.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.8 | 11.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.7 | 4.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.6 | 1.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.6 | 10.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 4.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 1.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.2 | 1.9 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 4.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.1 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.2 | 0.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 3.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 9.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 3.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 4.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 5.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 0.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 7.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 7.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 4.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 3.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 4.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 6.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0051424 | corticotropin-releasing hormone binding(GO:0051424) |
| 1.6 | 4.9 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.1 | 6.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.1 | 4.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.1 | 4.2 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 1.0 | 3.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.8 | 4.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.8 | 7.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 2.6 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.6 | 2.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.5 | 2.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.5 | 1.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.5 | 7.7 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.5 | 4.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 6.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 5.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.3 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 1.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 0.9 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.3 | 1.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 1.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 4.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 4.8 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 3.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 4.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 1.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 13.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 3.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 0.6 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.2 | 3.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 0.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.6 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 0.9 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 4.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 4.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 6.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 4.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 3.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 4.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.5 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.1 | 5.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 4.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 4.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 2.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 8.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 4.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.3 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 2.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 4.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 3.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 9.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 6.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 3.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 3.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 4.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 10.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 2.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 4.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 6.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 5.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 6.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.3 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 2.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |