Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
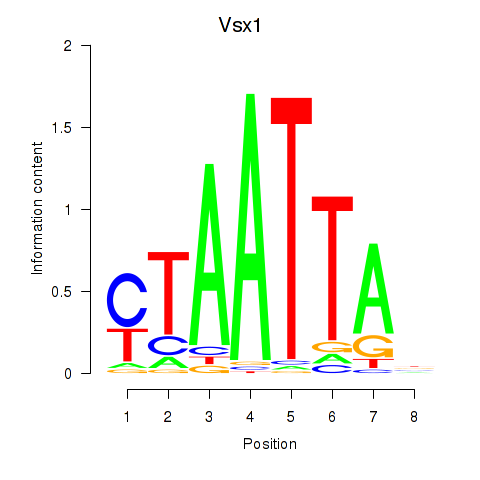
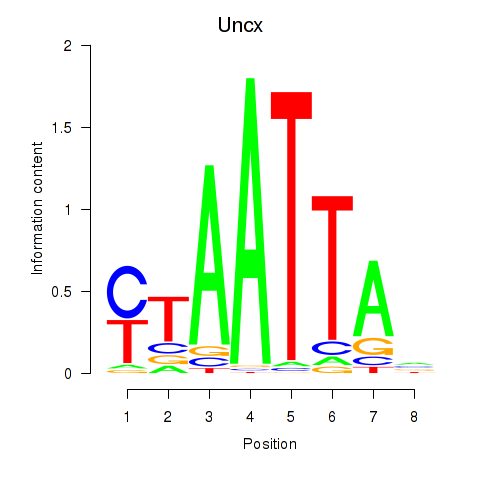
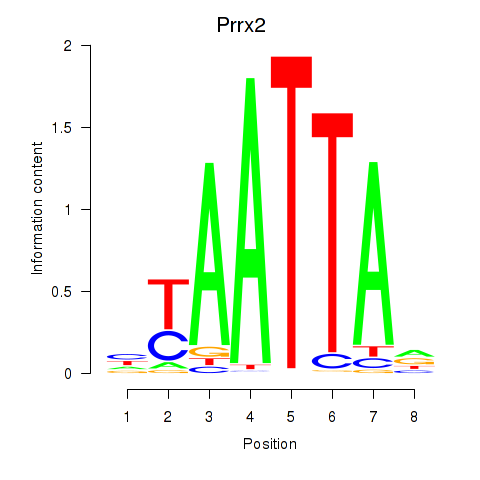
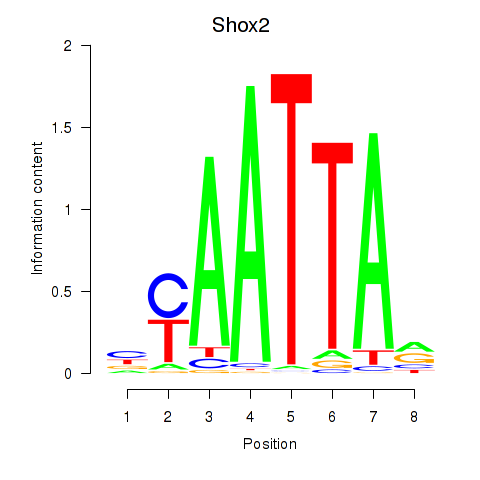
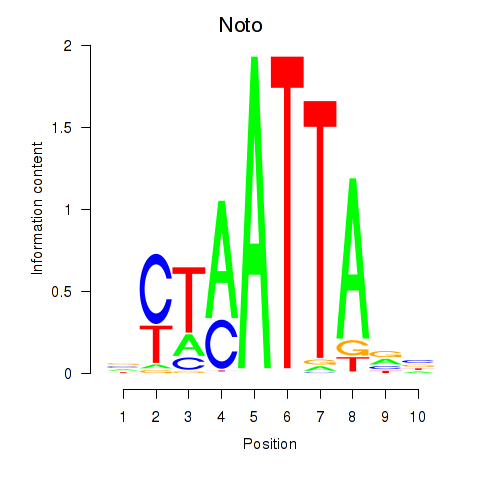
Results for Vsx1_Uncx_Prrx2_Shox2_Noto
Z-value: 0.91
Transcription factors associated with Vsx1_Uncx_Prrx2_Shox2_Noto
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vsx1
|
ENSMUSG00000033080.10 | Vsx1 |
|
Uncx
|
ENSMUSG00000029546.13 | Uncx |
|
Prrx2
|
ENSMUSG00000039476.14 | Prrx2 |
|
Shox2
|
ENSMUSG00000027833.17 | Shox2 |
|
Noto
|
ENSMUSG00000068302.9 | Noto |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prrx2 | mm39_v1_chr2_+_30724984_30724984 | -0.29 | 1.3e-02 | Click! |
| Noto | mm39_v1_chr6_+_85400868_85400868 | -0.28 | 1.6e-02 | Click! |
| Shox2 | mm39_v1_chr3_-_66888474_66888651 | 0.26 | 2.5e-02 | Click! |
| Uncx | mm39_v1_chr5_+_139529643_139529662 | 0.09 | 4.7e-01 | Click! |
| Vsx1 | mm39_v1_chr2_-_150531280_150531280 | 0.02 | 8.9e-01 | Click! |
Activity profile of Vsx1_Uncx_Prrx2_Shox2_Noto motif
Sorted Z-values of Vsx1_Uncx_Prrx2_Shox2_Noto motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx1_Uncx_Prrx2_Shox2_Noto
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_33662976 | 9.53 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr11_+_67090878 | 9.01 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr10_-_107321938 | 6.69 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr4_+_98919183 | 5.83 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr11_+_114741948 | 5.70 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_99213769 | 5.54 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr9_-_71070506 | 5.48 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr6_-_138056914 | 5.31 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chrX_+_132751729 | 5.20 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr6_-_50631418 | 5.12 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr11_+_114742331 | 4.99 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr4_-_35845204 | 4.94 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr3_+_82915031 | 4.53 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr18_-_43610829 | 4.51 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_+_114344920 | 4.00 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr13_-_113800172 | 3.97 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr1_-_163552693 | 3.76 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr10_-_107330580 | 3.67 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chrX_+_149330371 | 3.63 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr7_+_51528788 | 3.42 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr5_+_135135735 | 3.32 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chr6_-_41752111 | 3.24 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr10_+_50770836 | 3.21 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr1_+_53100796 | 3.18 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr2_-_168608949 | 3.06 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chrM_+_10167 | 3.01 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_+_55689921 | 3.00 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr11_-_100653754 | 2.96 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr1_-_72323464 | 2.90 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr10_-_76276278 | 2.80 |
ENSMUST00000235907.2
|
ENSMUSG00000117965.2
|
novel protein |
| chr6_-_138404076 | 2.72 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr2_-_34990689 | 2.72 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr1_-_72323407 | 2.70 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr7_-_4909515 | 2.67 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr2_+_124978612 | 2.67 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr2_+_124978518 | 2.66 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr14_-_64654397 | 2.64 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr11_+_67689094 | 2.62 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr9_+_118307250 | 2.61 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr19_-_7943365 | 2.57 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr17_-_84154173 | 2.53 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr11_-_99441687 | 2.52 |
ENSMUST00000092700.5
|
Krtap3-3
|
keratin associated protein 3-3 |
| chr3_+_59989282 | 2.52 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr16_+_22737227 | 2.48 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr15_-_34356567 | 2.45 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr5_+_104350475 | 2.44 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr6_-_30936013 | 2.43 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr14_-_64654592 | 2.41 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr16_+_22737128 | 2.36 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr17_-_84154196 | 2.35 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr5_+_88603847 | 2.34 |
ENSMUST00000198265.5
ENSMUST00000031226.9 |
Ambn
|
ameloblastin |
| chr16_+_22737050 | 2.34 |
ENSMUST00000231768.2
|
Fetub
|
fetuin beta |
| chr2_+_67935015 | 2.34 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr15_+_92495007 | 2.33 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr9_+_118307412 | 2.25 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr2_+_9887427 | 2.21 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr7_-_115423934 | 2.20 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_109835224 | 2.17 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr9_+_72714156 | 2.16 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr11_+_67061908 | 2.13 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr7_-_115459082 | 2.12 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_-_101765915 | 2.11 |
ENSMUST00000084830.2
|
Chrna10
|
cholinergic receptor, nicotinic, alpha polypeptide 10 |
| chr9_+_44309727 | 2.10 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr7_+_49559859 | 2.05 |
ENSMUST00000056442.12
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr8_-_3674993 | 2.05 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr7_-_44752508 | 2.05 |
ENSMUST00000209830.2
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr11_+_67061837 | 2.02 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr7_-_84339156 | 1.99 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr14_-_118289557 | 1.97 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr11_-_99265721 | 1.92 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr11_-_99494134 | 1.91 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr11_-_59466995 | 1.85 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr11_-_43792013 | 1.85 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr8_+_46080746 | 1.82 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_-_49286594 | 1.81 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr6_+_63232955 | 1.81 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr2_-_168607166 | 1.80 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chrX_-_142716085 | 1.76 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chrM_+_9870 | 1.76 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrX_+_99019176 | 1.75 |
ENSMUST00000113781.8
ENSMUST00000113783.8 ENSMUST00000113779.8 ENSMUST00000113776.8 ENSMUST00000113775.8 ENSMUST00000113780.8 ENSMUST00000113778.8 ENSMUST00000113777.8 ENSMUST00000071453.3 |
Eda
|
ectodysplasin-A |
| chr9_+_24194729 | 1.74 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr2_-_168609110 | 1.72 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr8_-_49008305 | 1.72 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr16_+_42727926 | 1.71 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr11_+_70410009 | 1.71 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr13_+_8252957 | 1.70 |
ENSMUST00000123187.2
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr11_+_70410445 | 1.70 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr12_-_11258973 | 1.70 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr1_-_9369463 | 1.69 |
ENSMUST00000140295.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr7_-_12829100 | 1.69 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr13_+_22563988 | 1.68 |
ENSMUST00000227685.2
ENSMUST00000227689.2 ENSMUST00000227846.2 |
Vmn1r199
|
vomeronasal 1 receptor 199 |
| chr14_-_109151590 | 1.67 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr7_-_44753168 | 1.66 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr1_-_9369045 | 1.65 |
ENSMUST00000191683.6
|
Sntg1
|
syntrophin, gamma 1 |
| chr8_+_84262409 | 1.64 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr2_-_27365633 | 1.64 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr19_+_39980868 | 1.63 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr5_+_108842294 | 1.63 |
ENSMUST00000013633.12
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr13_-_23041731 | 1.63 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr13_+_8252895 | 1.60 |
ENSMUST00000064473.13
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chrX_+_149372903 | 1.59 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr10_+_85763545 | 1.58 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr1_+_132155922 | 1.58 |
ENSMUST00000191418.2
|
Lemd1
|
LEM domain containing 1 |
| chr4_-_43823866 | 1.57 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr1_-_9368721 | 1.53 |
ENSMUST00000132064.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr4_-_97666279 | 1.52 |
ENSMUST00000146447.8
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr18_-_39000056 | 1.52 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr13_+_75855695 | 1.51 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr8_+_22996233 | 1.50 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr11_-_99228756 | 1.50 |
ENSMUST00000100482.3
|
Krt26
|
keratin 26 |
| chr19_-_13828056 | 1.50 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr7_+_126550009 | 1.49 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr15_-_34495329 | 1.48 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr13_+_22454692 | 1.48 |
ENSMUST00000228711.2
|
Vmn1r195
|
vomeronasal 1 receptor 195 |
| chr2_+_61634797 | 1.47 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr13_+_27241551 | 1.47 |
ENSMUST00000110369.10
ENSMUST00000224228.2 ENSMUST00000018061.7 |
Prl
|
prolactin |
| chr1_+_104696235 | 1.46 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr15_+_55171138 | 1.46 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr15_-_101710781 | 1.45 |
ENSMUST00000063292.8
|
Krt73
|
keratin 73 |
| chr19_-_13827773 | 1.45 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr1_+_88234454 | 1.44 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr11_-_99979052 | 1.43 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr3_-_154034271 | 1.43 |
ENSMUST00000204403.2
|
Lhx8
|
LIM homeobox protein 8 |
| chr14_+_54436247 | 1.42 |
ENSMUST00000103720.2
|
Traj21
|
T cell receptor alpha joining 21 |
| chrX_-_142716200 | 1.41 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr18_-_81029986 | 1.40 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr9_-_103099262 | 1.39 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr8_+_121842902 | 1.39 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr3_+_57332735 | 1.39 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr8_-_3675274 | 1.37 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr4_+_114914880 | 1.36 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr16_-_92196954 | 1.36 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr13_+_111391544 | 1.36 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr13_+_23991010 | 1.34 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr12_+_30934320 | 1.33 |
ENSMUST00000067087.7
|
Alkal2
|
ALK and LTK ligand 2 |
| chr9_-_44714263 | 1.32 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr7_+_126549859 | 1.32 |
ENSMUST00000106333.8
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr8_-_69541852 | 1.31 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr8_-_32408864 | 1.29 |
ENSMUST00000073884.7
ENSMUST00000238812.2 |
Nrg1
|
neuregulin 1 |
| chr8_+_46080840 | 1.28 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_+_83723743 | 1.28 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr5_+_139529643 | 1.28 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr7_-_101765970 | 1.27 |
ENSMUST00000211408.2
|
Chrna10
|
cholinergic receptor, nicotinic, alpha polypeptide 10 |
| chr9_+_38755745 | 1.26 |
ENSMUST00000217350.2
|
Olfr924
|
olfactory receptor 924 |
| chr12_+_38830812 | 1.25 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr8_-_3675024 | 1.24 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr6_+_37847721 | 1.23 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr9_+_37829844 | 1.23 |
ENSMUST00000212878.2
ENSMUST00000086061.4 |
Olfr878
|
olfactory receptor 878 |
| chr7_+_126549692 | 1.22 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr5_+_115373895 | 1.22 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr15_+_21111428 | 1.21 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr8_-_32408786 | 1.21 |
ENSMUST00000239000.2
|
Nrg1
|
neuregulin 1 |
| chr3_-_92441809 | 1.21 |
ENSMUST00000193521.2
|
2310046K23Rik
|
RIKEN cDNA 2310046K23 gene |
| chr9_-_37580478 | 1.20 |
ENSMUST00000011262.4
|
Panx3
|
pannexin 3 |
| chr2_+_87725306 | 1.19 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr1_-_189902868 | 1.18 |
ENSMUST00000177288.4
ENSMUST00000175916.8 |
Prox1
|
prospero homeobox 1 |
| chr19_-_45224251 | 1.17 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr2_+_69727563 | 1.16 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr18_+_23548192 | 1.15 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr11_-_107228382 | 1.15 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr3_+_76500857 | 1.14 |
ENSMUST00000162471.2
|
Fstl5
|
follistatin-like 5 |
| chr2_-_57942844 | 1.14 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr7_-_84339045 | 1.13 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr18_-_81029751 | 1.12 |
ENSMUST00000238808.2
|
Sall3
|
spalt like transcription factor 3 |
| chr15_+_39522905 | 1.11 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr15_-_11038077 | 1.11 |
ENSMUST00000058007.7
|
Rxfp3
|
relaxin family peptide receptor 3 |
| chr19_-_11838430 | 1.11 |
ENSMUST00000214796.2
|
Olfr1418
|
olfactory receptor 1418 |
| chr13_+_83723255 | 1.10 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_63014514 | 1.08 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr13_-_53627110 | 1.07 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr2_+_22959452 | 1.07 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr14_-_48902555 | 1.07 |
ENSMUST00000118578.9
|
Otx2
|
orthodenticle homeobox 2 |
| chr2_-_34951443 | 1.06 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr1_-_150341911 | 1.05 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr16_+_43574389 | 1.03 |
ENSMUST00000229953.2
|
Drd3
|
dopamine receptor D3 |
| chr11_+_116734104 | 1.03 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr13_-_23302396 | 1.02 |
ENSMUST00000227110.2
|
Vmn1r217
|
vomeronasal 1 receptor 217 |
| chr4_+_34893772 | 1.02 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr2_-_86857424 | 1.00 |
ENSMUST00000214857.2
ENSMUST00000215972.2 |
Olfr1104
|
olfactory receptor 1104 |
| chr15_+_101371353 | 1.00 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr1_-_14380418 | 1.00 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_71359000 | 1.00 |
ENSMUST00000126400.2
|
Dlx1
|
distal-less homeobox 1 |
| chr13_+_23214588 | 1.00 |
ENSMUST00000227652.2
ENSMUST00000227236.2 |
Vmn1r214
|
vomeronasal 1 receptor 214 |
| chr7_-_108774367 | 0.99 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr10_+_127734384 | 0.99 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr14_+_111912529 | 0.99 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chrX_+_151909893 | 0.98 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr9_-_76120939 | 0.97 |
ENSMUST00000074880.6
|
Gfral
|
GDNF family receptor alpha like |
| chr7_+_43418404 | 0.97 |
ENSMUST00000014063.6
|
Klk12
|
kallikrein related-peptidase 12 |
| chr18_-_3281089 | 0.97 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr17_+_38104420 | 0.96 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr1_-_92446383 | 0.96 |
ENSMUST00000062353.12
|
Olfr1414
|
olfactory receptor 1414 |
| chr13_-_23302027 | 0.95 |
ENSMUST00000228656.2
|
Vmn1r217
|
vomeronasal 1 receptor 217 |
| chr12_+_108145802 | 0.95 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr17_+_93506590 | 0.94 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr7_-_45480200 | 0.92 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr3_-_92031247 | 0.92 |
ENSMUST00000070284.4
|
Prr9
|
proline rich 9 |
| chr1_-_14380327 | 0.92 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_69727599 | 0.91 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr13_+_22474090 | 0.91 |
ENSMUST00000228382.2
ENSMUST00000228557.2 ENSMUST00000226245.2 ENSMUST00000227516.2 |
Vmn1r196
|
vomeronasal 1 receptor 196 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.4 | 5.5 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.3 | 10.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 1.2 | 4.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.1 | 3.2 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 1.0 | 5.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.9 | 1.7 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.8 | 3.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.7 | 2.0 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.6 | 3.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.6 | 1.8 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.5 | 4.9 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.5 | 2.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 2.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.5 | 5.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.5 | 1.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.5 | 1.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 1.3 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.4 | 4.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 1.7 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.4 | 0.9 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.4 | 1.7 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.4 | 1.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 3.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.4 | 1.1 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.4 | 1.8 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.3 | 1.4 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.3 | 1.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.3 | 1.0 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 4.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 3.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 2.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 0.9 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 3.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 2.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 14.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.3 | 6.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 2.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 1.5 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.2 | 2.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 2.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 4.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 2.0 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.2 | 3.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.2 | 0.6 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.2 | 1.6 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 2.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 2.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 3.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 2.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 0.5 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 1.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 3.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 1.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 3.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.8 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 2.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 3.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 4.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.4 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.5 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 0.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.4 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 7.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.5 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.3 | GO:0003166 | thigmotaxis(GO:0001966) bundle of His development(GO:0003166) |
| 0.1 | 1.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 4.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 3.1 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.3 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.6 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 1.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 3.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 13.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.7 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 5.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.8 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.7 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.2 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 1.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 7.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 2.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 9.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 1.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.7 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.4 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 1.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.6 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 37.0 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.6 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 1.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 5.0 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.8 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.9 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 3.3 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 3.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.3 | GO:1903859 | regulation of dendrite extension(GO:1903859) |
| 0.0 | 0.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 4.8 | GO:0015833 | peptide transport(GO:0015833) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 4.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.0 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.5 | 1.6 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.5 | 1.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.5 | 4.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 4.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 1.4 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.4 | 9.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 4.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 1.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 1.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 16.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 5.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 2.0 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 3.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 8.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 4.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 5.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.4 | 5.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.3 | 5.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.2 | 3.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.1 | 5.5 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.8 | 2.3 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.7 | 2.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.6 | 2.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 1.6 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.5 | 5.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.5 | 3.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 1.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.5 | 1.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 1.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.3 | 1.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 1.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 7.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 0.9 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 3.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 3.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.4 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.6 | GO:0071820 | N-box binding(GO:0071820) |
| 0.2 | 4.9 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 5.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 1.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 1.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 1.5 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 3.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 1.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 2.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 2.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 3.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 9.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 2.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 13.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 2.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.7 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 2.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 4.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 2.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 8.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 16.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) aromatase activity(GO:0070330) |
| 0.0 | 2.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 9.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 3.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 6.0 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 3.8 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 6.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 26.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 21.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 2.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 3.0 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 3.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 12.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 4.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 3.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 3.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 4.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 4.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 3.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 3.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 8.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 3.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 2.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |