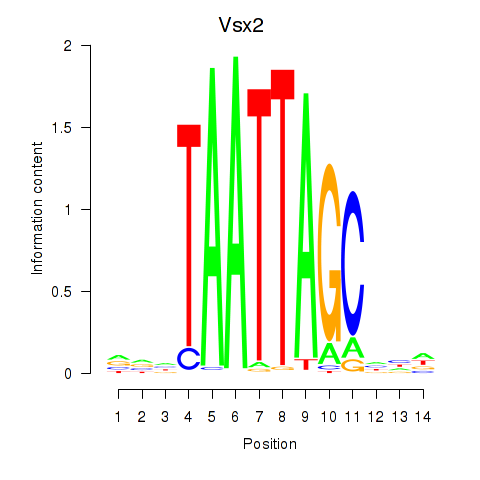
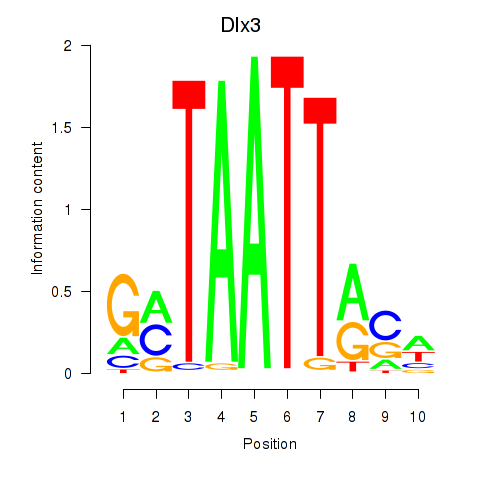
chr16_-_45544960
Show fit
4.40
ENSMUST00000096057.5
Tagln3
transgelin 3
chr18_+_57601541
Show fit
4.15
ENSMUST00000091892.4
ENSMUST00000209782.2
Ctxn3
cortexin 3
chr1_-_158183894
Show fit
3.14
ENSMUST00000004133.11
Brinp2
bone morphogenic protein/retinoic acid inducible neural-specific 2
chr1_-_79417732
Show fit
2.84
ENSMUST00000185234.2
ENSMUST00000049972.6
Scg2
secretogranin II
chr5_-_84565218
Show fit
2.80
ENSMUST00000113401.4
Epha5
Eph receptor A5
chr3_-_116047148
Show fit
2.28
ENSMUST00000090473.7
Gpr88
G-protein coupled receptor 88
chr3_-_80710097
Show fit
2.09
ENSMUST00000075316.10
ENSMUST00000107745.8
Gria2
glutamate receptor, ionotropic, AMPA2 (alpha 2)
chr18_+_23548534
Show fit
2.03
ENSMUST00000221880.2
ENSMUST00000220904.2
ENSMUST00000047954.15
Dtna
dystrobrevin alpha
chr5_-_70999547
Show fit
1.95
ENSMUST00000199705.2
Gabrg1
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1
chr10_+_73657689
Show fit
1.94
ENSMUST00000064562.14
ENSMUST00000193174.6
ENSMUST00000105426.10
ENSMUST00000129404.9
ENSMUST00000131321.9
ENSMUST00000126920.9
ENSMUST00000147189.9
ENSMUST00000105424.10
ENSMUST00000092420.13
ENSMUST00000105429.10
ENSMUST00000193361.6
ENSMUST00000131724.9
ENSMUST00000152655.9
ENSMUST00000151116.9
ENSMUST00000155701.9
ENSMUST00000152819.9
ENSMUST00000125517.9
ENSMUST00000124046.8
ENSMUST00000146682.8
ENSMUST00000177107.8
ENSMUST00000149977.9
ENSMUST00000191854.6
Pcdh15
protocadherin 15
chr14_+_99536111
Show fit
1.93
ENSMUST00000005279.8
Klf5
Kruppel-like factor 5
chr10_+_127734384
Show fit
1.82
ENSMUST00000047134.8
Sdr9c7
4short chain dehydrogenase/reductase family 9C, member 7
chr12_+_52746158
Show fit
1.81
ENSMUST00000095737.5
Akap6
A kinase (PRKA) anchor protein 6
chr16_-_34083549
Show fit
1.73
ENSMUST00000114949.8
ENSMUST00000114954.8
Kalrn
kalirin, RhoGEF kinase
chr2_+_84564394
Show fit
1.72
ENSMUST00000238573.2
ENSMUST00000090729.9
Ypel4
yippee like 4
chr1_+_172383499
Show fit
1.62
ENSMUST00000061835.10
Vsig8
V-set and immunoglobulin domain containing 8
chr18_+_23548455
Show fit
1.60
ENSMUST00000115832.4
Dtna
dystrobrevin alpha
chr12_+_29584560
Show fit
1.44
ENSMUST00000021009.10
Myt1l
myelin transcription factor 1-like
chr15_+_39522905
Show fit
1.42
ENSMUST00000226410.2
Rims2
regulating synaptic membrane exocytosis 2
chr1_-_134883645
Show fit
1.41
ENSMUST00000045665.13
ENSMUST00000086444.6
ENSMUST00000112163.2
Ppp1r12b
protein phosphatase 1, regulatory subunit 12B
chr4_+_133795740
Show fit
1.40
ENSMUST00000121391.8
Crybg2
crystallin beta-gamma domain containing 2
chr11_+_115197980
Show fit
1.40
ENSMUST00000055490.9
Otop2
otopetrin 2
chr12_-_31763859
Show fit
1.40
ENSMUST00000057783.6
ENSMUST00000236002.2
ENSMUST00000174480.3
ENSMUST00000176710.2
Gpr22
G protein-coupled receptor 22
chr5_+_27466914
Show fit
1.36
ENSMUST00000101471.4
Dpp6
dipeptidylpeptidase 6
chr2_-_79287095
Show fit
1.35
ENSMUST00000041099.5
Neurod1
neurogenic differentiation 1
chr6_+_77219698
Show fit
1.32
ENSMUST00000161677.2
Lrrtm1
leucine rich repeat transmembrane neuronal 1
chr6_+_77219627
Show fit
1.31
ENSMUST00000159616.2
Lrrtm1
leucine rich repeat transmembrane neuronal 1
chr18_+_23548192
Show fit
1.26
ENSMUST00000222515.2
Dtna
dystrobrevin alpha
chr17_+_17622934
Show fit
1.22
ENSMUST00000115576.3
Lix1
limb and CNS expressed 1
chr17_+_93506435
Show fit
1.18
ENSMUST00000234646.2
ENSMUST00000234081.2
Adcyap1
adenylate cyclase activating polypeptide 1
chr8_+_113296675
Show fit
1.18
ENSMUST00000034225.7
ENSMUST00000118171.8
Cntnap4
contactin associated protein-like 4
chr16_-_89615225
Show fit
1.18
ENSMUST00000164263.9
Tiam1
T cell lymphoma invasion and metastasis 1
chr7_+_126549692
Show fit
1.16
ENSMUST00000106335.8
ENSMUST00000146017.3
Sez6l2
seizure related 6 homolog like 2
chr17_+_93506590
Show fit
1.15
ENSMUST00000064775.8
Adcyap1
adenylate cyclase activating polypeptide 1
chr11_-_102469896
Show fit
1.12
ENSMUST00000107080.2
Gm11627
predicted gene 11627
chr3_-_130524024
Show fit
1.09
ENSMUST00000079085.11
Rpl34
ribosomal protein L34
chr1_-_134883577
Show fit
1.09
ENSMUST00000168381.8
Ppp1r12b
protein phosphatase 1, regulatory subunit 12B
chr9_-_71070506
Show fit
1.08
ENSMUST00000074465.9
Aqp9
aquaporin 9
chr6_-_102441628
Show fit
1.05
ENSMUST00000032159.7
Cntn3
contactin 3
chr2_+_177760959
Show fit
1.05
ENSMUST00000108916.8
Phactr3
phosphatase and actin regulator 3
chr6_+_63232955
Show fit
1.03
ENSMUST00000095852.5
Grid2
glutamate receptor, ionotropic, delta 2
chr10_-_85847697
Show fit
1.01
ENSMUST00000105304.2
ENSMUST00000061699.12
Bpifc
BPI fold containing family C
chr11_+_115225557
Show fit
1.00
ENSMUST00000106543.8
ENSMUST00000019006.5
Otop3
otopetrin 3
chr8_+_57774010
Show fit
0.95
ENSMUST00000040104.5
Hand2
Links
heart and neural crest derivatives expressed 2
chr3_+_55369149
Show fit
0.95
ENSMUST00000199585.5
ENSMUST00000070418.9
Dclk1
doublecortin-like kinase 1
chr3_-_130523954
Show fit
0.93
ENSMUST00000196202.5
ENSMUST00000133802.6
ENSMUST00000062601.14
ENSMUST00000200517.2
Rpl34
ribosomal protein L34
chr5_+_14075281
Show fit
0.92
ENSMUST00000073957.8
Sema3e
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
chr7_+_126550009
Show fit
0.92
ENSMUST00000106332.3
Sez6l2
seizure related 6 homolog like 2
chr15_-_37458768
Show fit
0.92
ENSMUST00000116445.9
Ncald
neurocalcin delta
chr3_-_50398027
Show fit
0.91
ENSMUST00000029297.6
ENSMUST00000194462.6
Slc7a11
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11
chr3_+_51568588
Show fit
0.91
ENSMUST00000099106.10
Mgst2
microsomal glutathione S-transferase 2
chr4_-_110149916
Show fit
0.89
ENSMUST00000106601.8
Elavl4
ELAV like RNA binding protein 4
chr6_+_30541581
Show fit
0.88
ENSMUST00000096066.5
Cpa2
carboxypeptidase A2, pancreatic
chr4_-_110143777
Show fit
0.84
ENSMUST00000138972.8
Elavl4
ELAV like RNA binding protein 4
chr2_-_32976378
Show fit
0.84
ENSMUST00000049618.9
Garnl3
GTPase activating RANGAP domain-like 3
chr13_+_83672654
Show fit
0.84
ENSMUST00000199019.5
Mef2c
myocyte enhancer factor 2C
chr10_+_59942274
Show fit
0.83
ENSMUST00000165024.3
Spock2
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2
chr6_+_106095726
Show fit
0.82
ENSMUST00000113258.8
ENSMUST00000079416.6
Cntn4
contactin 4
chr2_+_67935015
Show fit
0.82
ENSMUST00000042456.4
B3galt1
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
chr4_+_110254858
Show fit
0.80
ENSMUST00000106589.9
ENSMUST00000106587.9
ENSMUST00000106591.8
ENSMUST00000106592.8
Agbl4
ATP/GTP binding protein-like 4
chr9_-_119897358
Show fit
0.79
ENSMUST00000064165.5
Cx3cr1
chemokine (C-X3-C motif) receptor 1
chr3_-_151953894
Show fit
0.78
ENSMUST00000196529.5
Nexn
nexilin
chr7_-_115630282
Show fit
0.77
ENSMUST00000206034.2
ENSMUST00000106612.8
Sox6
SRY (sex determining region Y)-box 6
chrX_-_142716200
Show fit
0.76
ENSMUST00000112851.8
ENSMUST00000112856.3
ENSMUST00000033642.10
Dcx
doublecortin
chr3_+_5283606
Show fit
0.76
ENSMUST00000026284.13
Zfhx4
zinc finger homeodomain 4
chr14_-_36820304
Show fit
0.75
ENSMUST00000022337.11
Cdhr1
cadherin-related family member 1
chr12_+_38830081
Show fit
0.75
ENSMUST00000095767.11
Etv1
ets variant 1
chr9_-_119897328
Show fit
0.75
ENSMUST00000177637.2
Cx3cr1
chemokine (C-X3-C motif) receptor 1
chr6_-_136758716
Show fit
0.75
ENSMUST00000078095.11
ENSMUST00000032338.10
Gucy2c
guanylate cyclase 2c
chr17_+_35844091
Show fit
0.74
ENSMUST00000025273.9
Psors1c2
psoriasis susceptibility 1 candidate 2 (human)
chr19_-_47680528
Show fit
0.70
ENSMUST00000026045.14
ENSMUST00000086923.6
Col17a1
collagen, type XVII, alpha 1
chr6_-_147165623
Show fit
0.70
ENSMUST00000052296.9
ENSMUST00000204197.2
Pthlh
parathyroid hormone-like peptide
chr4_-_83203388
Show fit
0.67
ENSMUST00000150522.8
Ttc39b
tetratricopeptide repeat domain 39B
chr15_+_91722458
Show fit
0.66
ENSMUST00000109277.8
Smgc
submandibular gland protein C
chr8_-_3675274
Show fit
0.66
ENSMUST00000004749.7
Pcp2
Purkinje cell protein 2 (L7)
chr4_+_110254907
Show fit
0.64
ENSMUST00000097920.9
ENSMUST00000080744.13
Agbl4
ATP/GTP binding protein-like 4
chr12_+_99930757
Show fit
0.61
ENSMUST00000160413.8
ENSMUST00000177549.8
ENSMUST00000049788.9
Kcnk13
potassium channel, subfamily K, member 13
chr3_+_76500857
Show fit
0.61
ENSMUST00000162471.2
Fstl5
follistatin-like 5
chr11_-_102470287
Show fit
0.61
ENSMUST00000107081.8
Gm11627
predicted gene 11627
chr2_+_71359000
Show fit
0.59
ENSMUST00000126400.2
Dlx1
distal-less homeobox 1
chr5_-_143279378
Show fit
0.59
ENSMUST00000212715.2
Zfp853
zinc finger protein 853
chr7_+_126549859
Show fit
0.59
ENSMUST00000106333.8
Sez6l2
seizure related 6 homolog like 2
chr13_+_83672389
Show fit
0.58
ENSMUST00000200394.5
Mef2c
myocyte enhancer factor 2C
chr3_+_55689921
Show fit
0.58
ENSMUST00000075422.6
Mab21l1
mab-21-like 1
chr9_+_94551929
Show fit
0.57
ENSMUST00000033463.10
Slc9a9
solute carrier family 9 (sodium/hydrogen exchanger), member 9
chr10_-_43934774
Show fit
0.57
ENSMUST00000239010.2
Crybg1
crystallin beta-gamma domain containing 1
chr1_+_53100796
Show fit
0.56
ENSMUST00000027269.7
ENSMUST00000191197.2
Mstn
myostatin
chr15_+_91722524
Show fit
0.56
ENSMUST00000109276.8
ENSMUST00000088555.10
ENSMUST00000100293.9
ENSMUST00000126508.8
ENSMUST00000239545.1
Smgc
ENSMUSG00000118670.1
submandibular gland protein C
chr3_+_5283577
Show fit
0.55
ENSMUST00000175866.8
Zfhx4
zinc finger homeodomain 4
chr8_-_3675525
Show fit
0.53
ENSMUST00000144977.2
ENSMUST00000136105.8
ENSMUST00000128566.8
Pcp2
Purkinje cell protein 2 (L7)
chr9_+_32027335
Show fit
0.51
ENSMUST00000174641.8
Arhgap32
Rho GTPase activating protein 32
chr12_+_108572015
Show fit
0.51
ENSMUST00000109854.9
Evl
Ena-vasodilator stimulated phosphoprotein
chr11_-_101877832
Show fit
0.51
ENSMUST00000107173.9
ENSMUST00000107172.8
Dusp3
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related)
chr7_-_128020397
Show fit
0.50
ENSMUST00000147840.2
Rgs10
regulator of G-protein signalling 10
chr5_+_88731386
Show fit
0.50
ENSMUST00000031229.11
Rufy3
RUN and FYVE domain containing 3
chr8_-_3674993
Show fit
0.50
ENSMUST00000142431.8
Pcp2
Purkinje cell protein 2 (L7)
chr15_-_101602734
Show fit
0.50
ENSMUST00000023788.8
Krt6a
keratin 6A
chr13_+_118851214
Show fit
0.48
ENSMUST00000022246.9
Fgf10
fibroblast growth factor 10
chr11_+_102175985
Show fit
0.48
ENSMUST00000156326.2
Tmub2
transmembrane and ubiquitin-like domain containing 2
chr4_+_5724305
Show fit
0.48
ENSMUST00000108380.2
Fam110b
family with sequence similarity 110, member B
chr8_-_3675024
Show fit
0.48
ENSMUST00000133459.8
Pcp2
Purkinje cell protein 2 (L7)
chr2_+_31204314
Show fit
0.47
ENSMUST00000113532.9
Hmcn2
hemicentin 2
chr7_+_143792455
Show fit
0.46
ENSMUST00000239495.2
Shank2
SH3 and multiple ankyrin repeat domains 2
chr15_+_21111428
Show fit
0.46
ENSMUST00000075132.8
Cdh12
cadherin 12
chr12_+_38831093
Show fit
0.46
ENSMUST00000161513.9
Etv1
ets variant 1
chr5_+_88731366
Show fit
0.44
ENSMUST00000199312.5
Rufy3
RUN and FYVE domain containing 3
chr4_+_109835224
Show fit
0.42
ENSMUST00000061187.4
Dmrta2
doublesex and mab-3 related transcription factor like family A2
chr9_+_75682637
Show fit
0.41
ENSMUST00000012281.8
Bmp5
bone morphogenetic protein 5
chrX_-_142716085
Show fit
0.40
ENSMUST00000087313.10
Dcx
doublecortin
chr9_+_38738911
Show fit
0.38
ENSMUST00000051238.7
ENSMUST00000219798.2
Olfr923
olfactory receptor 923
chr17_+_44263890
Show fit
0.38
ENSMUST00000177857.9
ENSMUST00000044792.6
Rcan2
regulator of calcineurin 2
chr6_-_23248361
Show fit
0.38
ENSMUST00000031709.7
Fezf1
Fez family zinc finger 1
chr9_-_79885063
Show fit
0.37
ENSMUST00000093811.11
Filip1
filamin A interacting protein 1
chr14_+_10123804
Show fit
0.37
ENSMUST00000022262.6
ENSMUST00000224714.2
Fezf2
Fez family zinc finger 2
chr10_+_69623262
Show fit
0.37
ENSMUST00000183240.2
Ank3
ankyrin 3, epithelial
chr12_-_91815855
Show fit
0.35
ENSMUST00000167466.2
ENSMUST00000021347.12
ENSMUST00000178462.8
Sel1l
sel-1 suppressor of lin-12-like (C. elegans)
chr12_+_38830812
Show fit
0.35
ENSMUST00000160856.8
Etv1
ets variant 1
chr17_-_37409147
Show fit
0.35
ENSMUST00000216376.2
ENSMUST00000217372.2
Olfr91
olfactory receptor 91
chr10_-_44024843
Show fit
0.34
ENSMUST00000200401.2
Crybg1
crystallin beta-gamma domain containing 1
chr7_+_114344920
Show fit
0.34
ENSMUST00000136645.8
ENSMUST00000169913.8
Insc
INSC spindle orientation adaptor protein
chrX_+_132751729
Show fit
0.33
ENSMUST00000033602.9
Tnmd
tenomodulin
chr17_+_14087827
Show fit
0.33
ENSMUST00000239324.2
Afdn
afadin, adherens junction formation factor
chr19_-_42117420
Show fit
0.33
ENSMUST00000161873.2
ENSMUST00000018965.4
Avpi1
arginine vasopressin-induced 1
chr8_-_87307294
Show fit
0.32
ENSMUST00000131423.8
ENSMUST00000152438.2
Abcc12
ATP-binding cassette, sub-family C (CFTR/MRP), member 12
chr12_-_56660054
Show fit
0.32
ENSMUST00000072631.6
Nkx2-9
NK2 homeobox 9
chr7_-_115445352
Show fit
0.31
ENSMUST00000206369.2
Sox6
SRY (sex determining region Y)-box 6
chr9_-_79884920
Show fit
0.31
ENSMUST00000239133.2
Filip1
filamin A interacting protein 1
chr11_+_101877876
Show fit
0.30
ENSMUST00000010985.8
Cfap97d1
CFAP97 domain containing 1
chr1_-_163552693
Show fit
0.29
ENSMUST00000159679.8
Mettl11b
methyltransferase like 11B
chr9_+_24194729
Show fit
0.29
ENSMUST00000154644.2
Npsr1
neuropeptide S receptor 1
chr9_+_39784573
Show fit
0.28
ENSMUST00000079767.2
Olfr972
olfactory receptor 972
chr19_-_5610628
Show fit
0.28
ENSMUST00000025861.3
Ovol1
ovo like zinc finger 1
chrX_+_111003193
Show fit
0.27
ENSMUST00000130247.9
ENSMUST00000038546.7
Tex16
testis expressed gene 16
chr2_-_7400780
Show fit
0.26
ENSMUST00000002176.13
Celf2
CUGBP, Elav-like family member 2
chr6_-_30936013
Show fit
0.26
ENSMUST00000101589.5
Klf14
Kruppel-like factor 14
chr2_-_7400997
Show fit
0.26
ENSMUST00000137733.9
Celf2
CUGBP, Elav-like family member 2
chr7_-_102507962
Show fit
0.25
ENSMUST00000213481.2
ENSMUST00000209952.2
Olfr566
olfactory receptor 566
chr7_-_100504610
Show fit
0.25
ENSMUST00000156855.8
Relt
RELT tumor necrosis factor receptor
chr7_-_115445315
Show fit
0.24
ENSMUST00000166207.3
Sox6
SRY (sex determining region Y)-box 6
chr13_-_112698563
Show fit
0.23
ENSMUST00000224510.2
Il31ra
interleukin 31 receptor A
chr18_+_37433852
Show fit
0.23
ENSMUST00000051754.2
Pcdhb3
protocadherin beta 3
chr11_-_99228756
Show fit
0.22
ENSMUST00000100482.3
Krt26
keratin 26
chr17_-_37472385
Show fit
0.21
ENSMUST00000219235.3
Olfr93
olfactory receptor 93
chr15_-_101801351
Show fit
0.20
ENSMUST00000100179.2
Krt76
keratin 76
chr16_+_27126239
Show fit
0.20
ENSMUST00000066852.9
Ostn
osteocrin
chr2_+_49677688
Show fit
0.20
ENSMUST00000028103.13
Lypd6b
LY6/PLAUR domain containing 6B
chr4_+_108576846
Show fit
0.20
ENSMUST00000178992.2
3110021N24Rik
RIKEN cDNA 3110021N24 gene
chr18_+_37651393
Show fit
0.19
ENSMUST00000097609.3
Pcdhb22
protocadherin beta 22
chr10_+_127256192
Show fit
0.19
ENSMUST00000171434.8
R3hdm2
R3H domain containing 2
chr5_-_116427003
Show fit
0.19
ENSMUST00000086483.4
ENSMUST00000050178.13
Ccdc60
coiled-coil domain containing 60
chr2_-_7400690
Show fit
0.19
ENSMUST00000182404.8
Celf2
CUGBP, Elav-like family member 2
chr7_-_102566717
Show fit
0.19
ENSMUST00000214160.2
ENSMUST00000215773.2
Olfr571
olfactory receptor 571
chr9_-_79920131
Show fit
0.18
ENSMUST00000217264.2
Filip1
filamin A interacting protein 1
chr7_+_66489500
Show fit
0.18
ENSMUST00000107478.9
Adamts17
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17
chr17_+_21446349
Show fit
0.18
ENSMUST00000235895.2
Vmn1r234
vomeronasal 1 receptor 234
chr6_-_56546088
Show fit
0.18
ENSMUST00000203372.3
Pde1c
phosphodiesterase 1C
chr5_-_104225458
Show fit
0.18
ENSMUST00000198485.5
ENSMUST00000164471.8
ENSMUST00000178967.2
Gm17660
predicted gene, 17660
chr10_-_25076008
Show fit
0.18
ENSMUST00000100012.3
Akap7
A kinase (PRKA) anchor protein 7
chr2_+_89708781
Show fit
0.17
ENSMUST00000111519.3
Olfr1257
olfactory receptor 1257
chr7_+_106737534
Show fit
0.17
ENSMUST00000213367.3
ENSMUST00000214819.3
ENSMUST00000216871.3
ENSMUST00000215284.3
ENSMUST00000209942.2
Olfr716
olfactory receptor 716
chr10_+_128173603
Show fit
0.16
ENSMUST00000005826.9
Cs
citrate synthase
chr18_-_81029751
Show fit
0.15
ENSMUST00000238808.2
Sall3
spalt like transcription factor 3
chr8_-_32440071
Show fit
0.15
ENSMUST00000207678.3
Nrg1
neuregulin 1
chr18_-_38336893
Show fit
0.15
ENSMUST00000194312.2
Pcdh1
protocadherin 1
chr17_-_21216726
Show fit
0.15
ENSMUST00000237195.2
ENSMUST00000237629.2
ENSMUST00000056339.3
Vmn1r233
vomeronasal 1 receptor 233
chr8_+_21382681
Show fit
0.15
ENSMUST00000098908.4
Defb33
defensin beta 33
chr6_+_132739094
Show fit
0.14
ENSMUST00000069268.3
Tas2r102
taste receptor, type 2, member 102
chr2_-_168607166
Show fit
0.14
ENSMUST00000137536.2
Sall4
spalt like transcription factor 4
chr2_-_34990689
Show fit
0.14
ENSMUST00000226631.2
ENSMUST00000045776.5
ENSMUST00000226972.2
AI182371
expressed sequence AI182371
chr6_-_101354858
Show fit
0.13
ENSMUST00000075994.11
Pdzrn3
PDZ domain containing RING finger 3
chr17_-_37974666
Show fit
0.13
ENSMUST00000215414.2
ENSMUST00000213638.3
Olfr117
olfactory receptor 117
chr17_-_21006419
Show fit
0.13
ENSMUST00000233605.2
ENSMUST00000232812.2
ENSMUST00000233186.2
ENSMUST00000233164.2
Vmn1r228
vomeronasal 1 receptor 228
chr10_+_73657753
Show fit
0.12
ENSMUST00000134009.9
ENSMUST00000177420.7
ENSMUST00000125006.9
Pcdh15
protocadherin 15
chr15_-_34356567
Show fit
0.12
ENSMUST00000179647.2
9430069I07Rik
RIKEN cDNA 9430069I07 gene
chr7_+_66489465
Show fit
0.12
ENSMUST00000098382.10
Adamts17
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17
chr17_+_38110779
Show fit
0.12
ENSMUST00000215168.2
ENSMUST00000216478.2
Olfr124
olfactory receptor 124
chr6_-_68968278
Show fit
0.12
ENSMUST00000197966.2
Igkv4-81
immunoglobulin kappa variable 4-81
chr9_+_53678801
Show fit
0.11
ENSMUST00000048670.10
Slc35f2
solute carrier family 35, member F2
chr8_+_10299288
Show fit
0.11
ENSMUST00000214643.2
Myo16
myosin XVI
chr3_+_137770813
Show fit
0.11
ENSMUST00000163080.3
1110002E22Rik
RIKEN cDNA 1110002E22 gene
chr1_-_92446383
Show fit
0.10
ENSMUST00000062353.12
Olfr1414
olfactory receptor 1414
chr15_-_11038077
Show fit
0.10
ENSMUST00000058007.7
Rxfp3
relaxin family peptide receptor 3
chr4_-_154721288
Show fit
0.10
ENSMUST00000030902.13
ENSMUST00000105637.8
ENSMUST00000070313.14
ENSMUST00000105636.8
ENSMUST00000105638.9
ENSMUST00000097759.9
ENSMUST00000124771.2
Prdm16
PR domain containing 16
chr15_-_44291226
Show fit
0.10
ENSMUST00000227843.2
Nudcd1
NudC domain containing 1
chr13_-_103094784
Show fit
0.09
ENSMUST00000172264.8
ENSMUST00000099202.10
Mast4
microtubule associated serine/threonine kinase family member 4
chr11_-_99412162
Show fit
0.09
ENSMUST00000107445.8
Krt39
keratin 39
chr4_-_52919172
Show fit
0.09
ENSMUST00000107667.3
ENSMUST00000213989.2
Olfr272
olfactory receptor 272
chr12_-_111780268
Show fit
0.08
ENSMUST00000021715.6
Xrcc3
X-ray repair complementing defective repair in Chinese hamster cells 3
chr5_+_107656810
Show fit
0.08
ENSMUST00000160160.6
Gm42669
predicted gene 42669
chr14_-_52277310
Show fit
0.07
ENSMUST00000216907.2
ENSMUST00000214071.2
ENSMUST00000216188.2
Olfr221
olfactory receptor 221
chr10_-_8632519
Show fit
0.07
ENSMUST00000212869.2
Sash1
SAM and SH3 domain containing 1
chr11_-_99412084
Show fit
0.07
ENSMUST00000076948.2
Krt39
keratin 39
chr1_+_173983199
Show fit
0.07
ENSMUST00000213748.2
Olfr420
olfactory receptor 420
chr6_-_129752812
Show fit
0.07
ENSMUST00000095409.3
Klrh1
killer cell lectin-like receptor subfamily H, member 1
chr2_+_88644840
Show fit
0.06
ENSMUST00000214703.2
Olfr1202
olfactory receptor 1202
chr15_+_98329106
Show fit
0.06
ENSMUST00000215320.3
Olfr282
olfactory receptor 282
chr6_-_131662707
Show fit
0.06
ENSMUST00000072404.3
Tas2r104
taste receptor, type 2, member 104
chr9_+_20148415
Show fit
0.05
ENSMUST00000086474.6
Olfr872
olfactory receptor 872
chr2_-_90087936
Show fit
0.05
ENSMUST00000213968.2
Olfr142
olfactory receptor 142
chr1_-_179527845
Show fit
0.04
ENSMUST00000223392.2
Kif28
kinesin family member 28