Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Yy1_Yy2
Z-value: 1.81
Transcription factors associated with Yy1_Yy2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
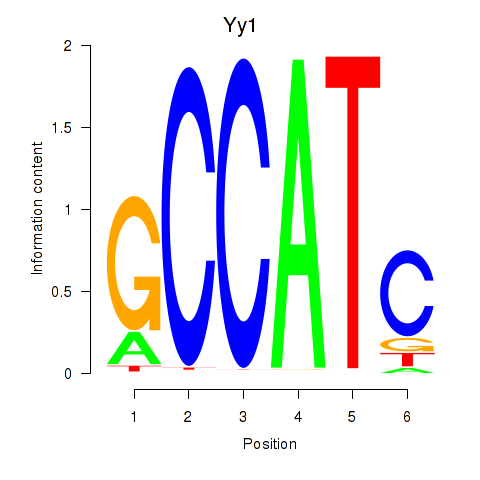
Yy1
|
ENSMUSG00000021264.13 | Yy1 |
|
Yy2
|
ENSMUSG00000091736.4 | Yy2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Yy2 | mm39_v1_chrX_-_156351979_156351987 | -0.25 | 3.8e-02 | Click! |
| Yy1 | mm39_v1_chr12_+_108758871_108758914 | -0.04 | 7.3e-01 | Click! |
Activity profile of Yy1_Yy2 motif
Sorted Z-values of Yy1_Yy2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Yy1_Yy2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_82572595 | 5.69 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr19_-_42074777 | 5.62 |
ENSMUST00000051772.10
|
Morn4
|
MORN repeat containing 4 |
| chr5_-_142594549 | 5.48 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr11_-_103844870 | 5.31 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr4_+_15881256 | 5.18 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chr2_+_49509288 | 5.13 |
ENSMUST00000028102.14
|
Kif5c
|
kinesin family member 5C |
| chr5_+_22951015 | 5.12 |
ENSMUST00000197992.2
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3 |
| chr15_+_87428483 | 5.02 |
ENSMUST00000230414.2
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr2_+_91757594 | 4.97 |
ENSMUST00000045537.4
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr2_+_25767795 | 4.91 |
ENSMUST00000114172.6
ENSMUST00000114176.9 ENSMUST00000037580.13 ENSMUST00000197917.5 |
Kcnt1
|
potassium channel, subfamily T, member 1 |
| chr5_-_144698443 | 4.88 |
ENSMUST00000061446.8
|
Tmem130
|
transmembrane protein 130 |
| chr17_+_26036893 | 4.85 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_174329361 | 4.72 |
ENSMUST00000030039.13
|
Fmn2
|
formin 2 |
| chr10_+_112292161 | 4.56 |
ENSMUST00000219607.2
ENSMUST00000218827.2 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr10_+_90412638 | 4.54 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_90412827 | 4.52 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_89338116 | 4.42 |
ENSMUST00000023291.6
|
Mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr16_+_17093941 | 4.31 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr2_-_92231598 | 4.22 |
ENSMUST00000050312.3
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr6_-_58884038 | 4.19 |
ENSMUST00000059539.5
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr7_+_46045862 | 4.07 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr5_+_137551774 | 4.04 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr19_+_8595369 | 3.99 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr16_-_94798509 | 3.82 |
ENSMUST00000095873.12
ENSMUST00000099508.4 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr14_-_76794103 | 3.77 |
ENSMUST00000064517.9
ENSMUST00000228055.2 |
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr10_+_90412570 | 3.73 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_136555364 | 3.68 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr15_+_83676140 | 3.65 |
ENSMUST00000172115.8
ENSMUST00000172398.2 |
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr1_+_158189831 | 3.60 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr8_-_122634418 | 3.57 |
ENSMUST00000045557.10
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr1_-_65162267 | 3.54 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr1_+_17215581 | 3.47 |
ENSMUST00000026879.8
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr9_-_20657643 | 3.45 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr9_+_110075133 | 3.45 |
ENSMUST00000199736.2
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr10_-_127098932 | 3.44 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr18_+_34973605 | 3.43 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr11_+_87651359 | 3.41 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr5_-_114952003 | 3.36 |
ENSMUST00000112160.4
|
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr7_+_27222678 | 3.34 |
ENSMUST00000108353.9
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr7_+_45434755 | 3.33 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chrX_+_20570145 | 3.31 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr15_+_4404965 | 3.30 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr17_-_24048069 | 3.28 |
ENSMUST00000069579.7
|
Elob
|
elongin B |
| chr7_+_44091822 | 3.24 |
ENSMUST00000058667.15
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr1_+_158190090 | 3.24 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr17_-_50600620 | 3.24 |
ENSMUST00000010736.9
|
Dazl
|
deleted in azoospermia-like |
| chr6_+_29468067 | 3.20 |
ENSMUST00000143101.4
ENSMUST00000149646.3 |
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr3_-_107366868 | 3.16 |
ENSMUST00000009617.10
ENSMUST00000238670.2 |
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr10_-_81066607 | 3.14 |
ENSMUST00000047408.6
|
Atcay
|
ataxia, cerebellar, Cayman type |
| chr5_+_137551790 | 3.14 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr13_-_99653045 | 3.12 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr5_-_38316296 | 3.11 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chrX_+_142447361 | 3.10 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr15_-_74713458 | 3.08 |
ENSMUST00000170259.3
|
Cyp11b1
|
cytochrome P450, family 11, subfamily b, polypeptide 1 |
| chr6_+_99669640 | 3.08 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr9_-_106343137 | 3.02 |
ENSMUST00000164834.3
|
Gpr62
|
G protein-coupled receptor 62 |
| chr18_-_67743854 | 3.02 |
ENSMUST00000115050.10
|
Spire1
|
spire type actin nucleation factor 1 |
| chr6_+_121277693 | 3.02 |
ENSMUST00000142419.2
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr18_-_43192483 | 3.01 |
ENSMUST00000025377.14
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_132294807 | 2.98 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr6_-_24956296 | 2.98 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr3_-_110051253 | 2.96 |
ENSMUST00000133268.9
ENSMUST00000051253.4 |
Ntng1
|
netrin G1 |
| chr18_-_25886908 | 2.95 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr16_+_20514925 | 2.94 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr8_+_70782467 | 2.92 |
ENSMUST00000207684.2
|
Gdf1
|
growth differentiation factor 1 |
| chr11_-_102360664 | 2.91 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr9_-_75591274 | 2.90 |
ENSMUST00000214244.2
ENSMUST00000213324.2 ENSMUST00000034699.8 |
Scg3
|
secretogranin III |
| chr10_+_90412432 | 2.90 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_38740784 | 2.90 |
ENSMUST00000226440.3
ENSMUST00000239553.1 |
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr11_+_16207705 | 2.89 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr8_+_31579633 | 2.89 |
ENSMUST00000170204.8
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr17_-_74017410 | 2.87 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr14_-_70864666 | 2.87 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chrX_+_134739783 | 2.87 |
ENSMUST00000173804.8
ENSMUST00000113136.8 |
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr2_-_32977182 | 2.85 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr11_-_75686874 | 2.85 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr3_-_107425316 | 2.84 |
ENSMUST00000169449.8
ENSMUST00000029499.15 |
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr14_+_68321302 | 2.83 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr4_-_126647156 | 2.83 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chr2_-_25209107 | 2.82 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr11_-_69451012 | 2.82 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr5_-_38316706 | 2.81 |
ENSMUST00000201341.2
ENSMUST00000201363.4 ENSMUST00000201134.2 |
Nsg1
|
neuron specific gene family member 1 |
| chr19_+_8814782 | 2.81 |
ENSMUST00000171649.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_-_35383867 | 2.80 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr9_-_54569128 | 2.80 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr6_-_124745294 | 2.80 |
ENSMUST00000135626.8
|
Eno2
|
enolase 2, gamma neuronal |
| chr8_-_125296435 | 2.80 |
ENSMUST00000238882.2
ENSMUST00000063278.7 |
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr11_+_108286114 | 2.76 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr2_-_25359752 | 2.76 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr18_-_43032535 | 2.75 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_+_44145987 | 2.74 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr13_+_75237939 | 2.73 |
ENSMUST00000022075.6
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr6_+_48963795 | 2.71 |
ENSMUST00000037696.6
|
Svs1
|
seminal vesicle secretory protein 1 |
| chr17_-_37334240 | 2.71 |
ENSMUST00000102665.11
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr1_+_34840785 | 2.70 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr4_+_152423075 | 2.69 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr9_+_89791943 | 2.69 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr3_-_72964276 | 2.68 |
ENSMUST00000192477.2
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr17_+_46421908 | 2.66 |
ENSMUST00000024763.10
ENSMUST00000123646.2 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr1_+_129201081 | 2.65 |
ENSMUST00000073527.13
ENSMUST00000040311.14 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr2_+_28095660 | 2.65 |
ENSMUST00000102879.4
ENSMUST00000028177.11 |
Olfm1
|
olfactomedin 1 |
| chr6_-_28831746 | 2.65 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr1_+_66403774 | 2.63 |
ENSMUST00000077355.12
ENSMUST00000114012.8 |
Map2
|
microtubule-associated protein 2 |
| chr11_+_58221538 | 2.63 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr8_+_31579499 | 2.63 |
ENSMUST00000036631.14
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr11_-_58346806 | 2.62 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr15_-_74728088 | 2.62 |
ENSMUST00000238900.2
ENSMUST00000167634.2 |
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr4_-_124755964 | 2.62 |
ENSMUST00000064444.8
|
Maneal
|
mannosidase, endo-alpha-like |
| chr17_-_24752683 | 2.61 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr7_-_5128936 | 2.61 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr10_+_7465555 | 2.60 |
ENSMUST00000134346.8
ENSMUST00000019931.12 ENSMUST00000130590.8 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr19_-_4956626 | 2.60 |
ENSMUST00000237085.2
ENSMUST00000053506.8 ENSMUST00000238170.2 |
Bbs1
|
Bardet-Biedl syndrome 1 (human) |
| chr8_-_72124359 | 2.60 |
ENSMUST00000177517.8
ENSMUST00000030170.15 |
Unc13a
|
unc-13 homolog A |
| chr7_+_44146012 | 2.59 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr1_-_75240551 | 2.59 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr2_+_164810139 | 2.56 |
ENSMUST00000202479.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr7_-_46783432 | 2.56 |
ENSMUST00000102626.10
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr8_+_85141975 | 2.55 |
ENSMUST00000121390.8
ENSMUST00000122053.2 |
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr10_-_70491764 | 2.54 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr6_-_124410452 | 2.53 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr3_+_58913234 | 2.50 |
ENSMUST00000040846.15
|
Med12l
|
mediator complex subunit 12-like |
| chr4_+_134070994 | 2.50 |
ENSMUST00000081094.6
|
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr16_+_34842764 | 2.50 |
ENSMUST00000061156.10
|
Hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr1_-_172156828 | 2.48 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr2_+_148237258 | 2.46 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr5_+_34153328 | 2.45 |
ENSMUST00000056355.9
|
Nat8l
|
N-acetyltransferase 8-like |
| chr13_-_36301466 | 2.45 |
ENSMUST00000053265.8
|
Lyrm4
|
LYR motif containing 4 |
| chr14_-_9015639 | 2.44 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr16_+_39804711 | 2.44 |
ENSMUST00000187695.7
|
Lsamp
|
limbic system-associated membrane protein |
| chr19_+_42235983 | 2.43 |
ENSMUST00000122375.8
|
Golga7b
|
golgi autoantigen, golgin subfamily a, 7B |
| chr8_+_112737997 | 2.42 |
ENSMUST00000052138.11
|
Terf2ip
|
telomeric repeat binding factor 2, interacting protein |
| chr2_-_25360043 | 2.39 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr4_+_110254858 | 2.39 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr7_-_109380745 | 2.39 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chr15_-_95426419 | 2.39 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr17_+_69144053 | 2.39 |
ENSMUST00000178545.3
|
Tmem200c
|
transmembrane protein 200C |
| chr17_-_37334091 | 2.38 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr2_-_156833932 | 2.37 |
ENSMUST00000109558.2
ENSMUST00000069600.13 ENSMUST00000072298.13 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr7_-_118047394 | 2.37 |
ENSMUST00000203796.4
ENSMUST00000203485.3 |
Syt17
|
synaptotagmin XVII |
| chr19_+_46750016 | 2.37 |
ENSMUST00000099373.12
ENSMUST00000077666.6 |
Cnnm2
|
cyclin M2 |
| chr11_-_82655132 | 2.36 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chrX_+_98864627 | 2.36 |
ENSMUST00000096363.3
|
Tmem28
|
transmembrane protein 28 |
| chr4_-_148123223 | 2.35 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr3_-_51304137 | 2.34 |
ENSMUST00000141156.3
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr10_+_90412539 | 2.34 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_-_98705791 | 2.33 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr4_+_110254907 | 2.33 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr9_-_8134295 | 2.33 |
ENSMUST00000037397.8
|
Cep126
|
centrosomal protein 126 |
| chr11_+_58221569 | 2.32 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr16_-_16687119 | 2.32 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr7_-_101513300 | 2.32 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr2_+_96148418 | 2.32 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr18_-_32271224 | 2.31 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chrX_-_101950075 | 2.31 |
ENSMUST00000120808.8
ENSMUST00000121197.8 |
Dmrtc1a
|
DMRT-like family C1a |
| chr1_-_175319842 | 2.31 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr14_-_70864448 | 2.31 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr3_-_51303924 | 2.30 |
ENSMUST00000108046.8
ENSMUST00000038154.12 |
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr1_+_158189900 | 2.29 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr1_-_135934080 | 2.29 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr8_+_110292524 | 2.29 |
ENSMUST00000034159.8
|
Txnl4b
|
thioredoxin-like 4B |
| chr15_+_99499252 | 2.28 |
ENSMUST00000230075.2
ENSMUST00000023754.6 |
Aqp6
|
aquaporin 6 |
| chr5_+_31652079 | 2.28 |
ENSMUST00000076949.13
ENSMUST00000202394.4 |
Gpn1
|
GPN-loop GTPase 1 |
| chr7_-_140402037 | 2.28 |
ENSMUST00000106052.2
ENSMUST00000080651.13 |
Zfp941
|
zinc finger protein 941 |
| chr2_+_29779721 | 2.27 |
ENSMUST00000113767.8
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr10_-_70435114 | 2.27 |
ENSMUST00000046513.10
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr3_-_84128160 | 2.27 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr7_+_92729067 | 2.26 |
ENSMUST00000051179.12
|
Fam181b
|
family with sequence similarity 181, member B |
| chr2_-_73605387 | 2.26 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr1_+_60448813 | 2.26 |
ENSMUST00000188594.7
ENSMUST00000188618.7 ENSMUST00000189980.7 |
Abi2
|
abl interactor 2 |
| chr12_-_109034099 | 2.25 |
ENSMUST00000190647.3
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr4_+_42922253 | 2.25 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24 |
| chr10_+_34173426 | 2.25 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chrX_+_134643435 | 2.25 |
ENSMUST00000096321.3
ENSMUST00000113144.8 ENSMUST00000113147.8 ENSMUST00000113145.8 |
Armcx5
Gprasp1
|
armadillo repeat containing, X-linked 5 G protein-coupled receptor associated sorting protein 1 |
| chr6_+_79995860 | 2.25 |
ENSMUST00000147663.8
ENSMUST00000128718.8 ENSMUST00000126005.8 ENSMUST00000133918.8 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr6_+_122851433 | 2.25 |
ENSMUST00000032477.6
|
Necap1
|
NECAP endocytosis associated 1 |
| chr7_-_103778992 | 2.24 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr14_-_4198510 | 2.24 |
ENSMUST00000133460.2
ENSMUST00000132374.9 |
Nkiras1
|
NFKB inhibitor interacting Ras-like protein 1 |
| chr16_-_4831349 | 2.24 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr2_-_73605684 | 2.23 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr16_-_34083549 | 2.22 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr16_+_41353360 | 2.21 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr7_-_79267390 | 2.20 |
ENSMUST00000032766.5
|
Rhcg
|
Rhesus blood group-associated C glycoprotein |
| chr3_+_114874614 | 2.19 |
ENSMUST00000051309.9
|
Olfm3
|
olfactomedin 3 |
| chr8_-_125161061 | 2.18 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr17_+_35455532 | 2.18 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_+_78432867 | 2.18 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr13_-_55563028 | 2.18 |
ENSMUST00000054146.5
|
Pfn3
|
profilin 3 |
| chr10_+_81068980 | 2.17 |
ENSMUST00000144087.2
ENSMUST00000117798.8 |
Zfr2
|
zinc finger RNA binding protein 2 |
| chr6_+_124973644 | 2.17 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr1_+_153528689 | 2.17 |
ENSMUST00000041776.12
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr16_-_34334314 | 2.17 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr18_-_36899245 | 2.17 |
ENSMUST00000061522.8
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr5_-_120641658 | 2.16 |
ENSMUST00000031597.7
|
Plbd2
|
phospholipase B domain containing 2 |
| chr11_+_102284229 | 2.16 |
ENSMUST00000107105.9
ENSMUST00000107102.8 ENSMUST00000107103.8 ENSMUST00000006750.8 |
Rundc3a
|
RUN domain containing 3A |
| chr11_-_69553451 | 2.16 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr7_+_27967222 | 2.16 |
ENSMUST00000059596.8
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr19_+_5012362 | 2.15 |
ENSMUST00000236917.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr15_+_80139371 | 2.15 |
ENSMUST00000109605.5
ENSMUST00000229828.2 |
Atf4
|
activating transcription factor 4 |
| chr1_+_183078573 | 2.14 |
ENSMUST00000109166.8
|
Aida
|
axin interactor, dorsalization associated |
| chr5_+_16139683 | 2.14 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 1.9 | 7.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 1.9 | 5.7 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 1.8 | 19.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.5 | 6.1 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.5 | 4.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.5 | 6.0 | GO:0021586 | pons maturation(GO:0021586) |
| 1.5 | 4.5 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.4 | 2.9 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 1.4 | 5.7 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.4 | 4.1 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 1.3 | 5.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 1.3 | 7.5 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.2 | 3.7 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.1 | 5.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.1 | 18.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.1 | 4.4 | GO:0021750 | vestibular nucleus development(GO:0021750) |
| 1.1 | 6.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.1 | 5.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.1 | 1.1 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 1.1 | 3.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.1 | 3.2 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 1.0 | 3.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.0 | 15.7 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 3.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.0 | 5.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.0 | 3.0 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 1.0 | 4.9 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 1.0 | 3.0 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 1.0 | 2.9 | GO:0018003 | peptidyl-lysine N6-acetylation(GO:0018003) |
| 1.0 | 5.8 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 1.0 | 1.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 1.0 | 8.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.9 | 4.7 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.9 | 10.9 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.9 | 1.8 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.9 | 5.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.9 | 2.6 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.8 | 0.8 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.8 | 2.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.8 | 2.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 3.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.8 | 3.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.8 | 2.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 | 5.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.8 | 2.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.8 | 2.3 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.8 | 3.0 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.7 | 3.6 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.7 | 2.9 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.7 | 2.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.7 | 2.8 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.7 | 6.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.7 | 2.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.7 | 4.9 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.7 | 12.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.7 | 3.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.7 | 4.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.7 | 2.0 | GO:0032499 | detection of peptidoglycan(GO:0032499) |
| 0.7 | 2.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.7 | 2.6 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.7 | 3.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.6 | 5.7 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.6 | 1.3 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.6 | 1.9 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.6 | 2.5 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.6 | 1.8 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.6 | 1.8 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 1.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.6 | 2.4 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.6 | 7.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.6 | 1.8 | GO:2000487 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 0.6 | 4.1 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.6 | 1.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.6 | 1.2 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.6 | 4.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 | 4.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.6 | 15.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.6 | 2.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.6 | 6.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 | 1.1 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.6 | 1.7 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.5 | 1.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.5 | 2.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.5 | 1.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.5 | 6.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.5 | 2.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.5 | 2.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.5 | 5.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.5 | 2.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 2.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.5 | 1.6 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.5 | 4.6 | GO:0032328 | alanine transport(GO:0032328) |
| 0.5 | 0.5 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.5 | 2.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.5 | 8.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.5 | 0.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.5 | 1.5 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.5 | 0.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.5 | 2.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 3.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.5 | 0.5 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.5 | 2.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.5 | 2.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.5 | 2.9 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.5 | 3.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.5 | 1.5 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.5 | 0.5 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.5 | 3.9 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.5 | 3.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 2.4 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.5 | 1.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.5 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.5 | 1.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 0.5 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.5 | 2.4 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.5 | 1.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.5 | 1.9 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.5 | 5.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 3.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 3.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 3.6 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.4 | 2.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 1.3 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.4 | 1.8 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 1.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.4 | 1.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.4 | 2.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 5.6 | GO:0097033 | mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 | 0.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.4 | 5.9 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.4 | 0.4 | GO:1902992 | negative regulation of amyloid precursor protein catabolic process(GO:1902992) |
| 0.4 | 0.4 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.4 | 1.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 1.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 2.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.4 | 15.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.4 | 2.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.4 | 1.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 2.0 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.4 | 1.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.4 | 14.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 1.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.4 | 1.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 1.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 1.6 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 1.2 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.4 | 3.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 | 0.4 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.4 | 1.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.4 | 1.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.4 | 2.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 3.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 1.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 2.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 1.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 | 2.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 2.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 5.9 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.3 | 1.7 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.3 | 1.4 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.3 | 1.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.3 | 1.7 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 1.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.3 | 1.4 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 2.0 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.3 | 7.1 | GO:0031498 | chromatin disassembly(GO:0031498) |
| 0.3 | 2.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 | 1.0 | GO:0072248 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.3 | 1.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 1.7 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.3 | 1.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 3.0 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 1.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 1.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.6 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.3 | 1.3 | GO:0015755 | fructose transport(GO:0015755) |
| 0.3 | 1.0 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.3 | 1.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 1.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 1.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 4.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.3 | 1.6 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.3 | 1.6 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 5.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.3 | 0.3 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.3 | 0.9 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.3 | 0.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.3 | 2.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 2.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.3 | 1.5 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.3 | 4.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 1.5 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.3 | 1.8 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.3 | 3.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 0.6 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.3 | 7.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 1.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 1.2 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.3 | 0.6 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.3 | 1.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 2.9 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 0.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.3 | 0.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 2.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 0.8 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.7 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 0.8 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.3 | 1.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 | 0.8 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.3 | 2.5 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.3 | 1.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 3.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 1.4 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.3 | 0.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 0.8 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.3 | 1.6 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.3 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 1.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.3 | 0.8 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.3 | 1.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 0.8 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 | 2.4 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.3 | 0.5 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 1.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 1.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.3 | 2.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 1.3 | GO:0048686 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 0.8 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 0.8 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.3 | 0.5 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 1.6 | GO:0090212 | negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.3 | 0.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 0.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.3 | 1.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 2.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 1.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.3 | 0.8 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 0.8 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.3 | 0.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.3 | 2.8 | GO:0015879 | amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.3 | 7.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 0.8 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.3 | 1.3 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.3 | 4.8 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.3 | 1.0 | GO:0044704 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.3 | 1.8 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 0.8 | GO:0007228 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.3 | 4.0 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 1.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 2.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.7 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 2.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.7 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 2.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 0.5 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 1.0 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 | 11.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 0.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.5 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 4.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 4.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 1.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.7 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 11.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.9 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.2 | 1.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.2 | 0.2 | GO:0070666 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.2 | 1.4 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.2 | 0.5 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 1.8 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 14.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 1.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 1.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 1.4 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.2 | 1.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 0.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 0.4 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.2 | 1.8 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.4 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 1.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 0.2 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.2 | 2.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 3.7 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.2 | 1.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 1.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 3.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 0.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 2.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.5 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 0.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 1.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 2.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.6 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.2 | 0.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 1.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.8 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.2 | 1.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 1.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 0.8 | GO:0009750 | response to fructose(GO:0009750) |
| 0.2 | 0.8 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body catabolic process(GO:0046952) |
| 0.2 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 0.8 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 0.4 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 0.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 1.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 0.6 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 1.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.8 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 1.0 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 3.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.4 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.2 | 5.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 0.8 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.2 | 1.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 1.2 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 0.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 1.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 1.6 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 | 0.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.2 | 1.0 | GO:1904181 | positive regulation of membrane depolarization(GO:1904181) |
| 0.2 | 0.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 4.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 1.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 0.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.2 | 7.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 2.7 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 0.8 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.2 | 0.6 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 1.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 2.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.2 | 1.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.2 | 3.8 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 0.6 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 2.5 | GO:0007135 | meiosis II(GO:0007135) |
| 0.2 | 1.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 0.6 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.2 | 1.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 0.2 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.4 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.2 | 0.4 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.2 | 0.8 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 0.4 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.2 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 | 0.6 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 1.7 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.2 | 1.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.2 | 6.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 3.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 0.2 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.2 | 0.7 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 1.1 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 2.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 1.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 0.5 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.2 | 1.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 2.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 2.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 0.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 2.7 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.2 | 0.7 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 2.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 3.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 0.9 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 2.1 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.2 | 2.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 1.4 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 0.7 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.2 | 1.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 1.0 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.2 | 2.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.5 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 2.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 0.3 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 0.3 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.2 | 1.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 0.3 | GO:0019230 | proprioception(GO:0019230) |
| 0.2 | 3.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 0.5 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 0.2 | 0.5 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.2 | 0.7 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.2 | 1.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 0.7 | GO:0008306 | associative learning(GO:0008306) |
| 0.2 | 1.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 5.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 4.0 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 1.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 4.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.2 | 0.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 0.3 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 0.6 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.2 | 6.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 0.5 | GO:0045851 | pH reduction(GO:0045851) |
| 0.2 | 0.5 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.2 | 0.8 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.2 | 0.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 4.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.2 | 2.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.3 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.2 | 1.2 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 3.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.2 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.2 | 0.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 3.7 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.2 | 1.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 2.6 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 0.2 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 1.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 0.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.5 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 4.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.2 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 1.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 0.3 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 1.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 1.0 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 1.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 4.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.6 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 5.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.4 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.9 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.7 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 1.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 3.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.4 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.7 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.1 | 1.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 1.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.4 | GO:0099630 | postsynaptic neurotransmitter receptor cycle(GO:0099630) |
| 0.1 | 0.6 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.1 | 0.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.6 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 1.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.6 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 3.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.6 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 2.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 1.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 3.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 5.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 1.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 3.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 4.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 1.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.4 | GO:0045141 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 1.6 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.1 | 0.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 0.7 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.8 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.7 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 1.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 3.1 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 3.3 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 0.5 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 1.5 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.5 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.8 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 0.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 3.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 8.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.6 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 1.1 | GO:0090186 | regulation of pancreatic juice secretion(GO:0090186) negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.1 | 0.7 | GO:0051324 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.1 | 2.1 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.2 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 3.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 1.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.6 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.5 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 3.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.6 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.1 | 0.5 | GO:0008050 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.8 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.8 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 1.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.5 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 0.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.8 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 0.3 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 1.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 4.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.5 | GO:0006586 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) |
| 0.1 | 1.7 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.2 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 2.0 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.5 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.1 | 0.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 1.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 0.3 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.2 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.1 | 2.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 2.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.1 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 1.9 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.1 | 1.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.9 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 2.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.3 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 0.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 2.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 0.4 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 1.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.6 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.7 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.1 | 1.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.4 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 4.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.9 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 2.0 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.3 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 3.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 1.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.6 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.2 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.1 | 1.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.3 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 2.6 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.9 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 3.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.0 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.3 | GO:0035854 | basophil differentiation(GO:0030221) eosinophil fate commitment(GO:0035854) |
| 0.1 | 1.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.2 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.1 | 0.5 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.1 | 0.3 | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) |
| 0.1 | 1.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.2 | GO:0046036 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.1 | 0.2 | GO:0002191 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial RNA 3'-end processing(GO:0000965) mitochondrial tRNA processing(GO:0090646) mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.4 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.1 | 1.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 10.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 2.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.1 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.6 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.0 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 | 1.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.2 | GO:0097276 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.1 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.6 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.7 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.9 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 6.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 2.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.5 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.1 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 2.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.1 | 1.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.9 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.9 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 0.4 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 0.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.1 | GO:0030815 | negative regulation of cAMP metabolic process(GO:0030815) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.4 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.7 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.2 | GO:0097286 | iron ion import(GO:0097286) |
| 0.1 | 4.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 2.0 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.8 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.5 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 1.9 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.2 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.1 | 0.8 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 14.8 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.4 | GO:2000109 | macrophage apoptotic process(GO:0071888) regulation of macrophage apoptotic process(GO:2000109) |
| 0.1 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.2 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:0090042 | tubulin deacetylation(GO:0090042) |
| 0.1 | 0.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.5 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.2 | GO:0050932 | regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 2.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 | 0.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.9 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.3 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 1.3 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.3 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 6.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 41.0 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0071372 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 1.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.1 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 1.0 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.4 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 2.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 3.6 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 3.7 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 2.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.0 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.3 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.3 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.8 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.1 | GO:0072199 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0071931 | trophectodermal cell proliferation(GO:0001834) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.4 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0009107 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 1.9 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 3.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 9.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 3.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.8 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0046112 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) nucleobase biosynthetic process(GO:0046112) |
| 0.0 | 0.5 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.9 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.0 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 | 0.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.0 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.0 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0030817 | cAMP biosynthetic process(GO:0006171) regulation of cAMP biosynthetic process(GO:0030817) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.1 | 6.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.1 | 10.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.0 | 4.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.0 | 4.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 1.0 | 5.9 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.9 | 6.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.9 | 0.9 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.8 | 3.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 3.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.8 | 0.8 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.8 | 9.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.8 | 3.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.8 | 6.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.7 | 11.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 2.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.6 | 1.8 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.6 | 1.8 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.6 | 3.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.6 | 17.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.6 | 4.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 4.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 7.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.6 | 1.7 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.6 | 1.7 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.5 | 2.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 3.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 7.7 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.5 | 6.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.5 | 22.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.5 | 1.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.5 | 11.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 23.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 5.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 1.8 | GO:0071920 | cleavage body(GO:0071920) |
| 0.4 | 1.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 11.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 0.4 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.4 | 14.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 16.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 1.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 1.2 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.4 | 10.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 7.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.4 | 2.6 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 5.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 12.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 4.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 5.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 3.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 1.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 1.8 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.4 | 1.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 2.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 1.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 1.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.3 | 5.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 3.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.3 | 1.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 2.9 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.3 | 1.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 0.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 1.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 4.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 0.9 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.3 | 1.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 1.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 2.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.3 | 4.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 6.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 0.8 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.3 | 1.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 1.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 1.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 2.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 0.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 0.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 13.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 0.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 2.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.5 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 0.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 3.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 3.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.8 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 10.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 0.4 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.2 | 10.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 2.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 0.7 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 0.7 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 0.4 | GO:0044317 | rod spherule(GO:0044317) |
| 0.2 | 2.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 17.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 69.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 1.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 2.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.2 | 5.6 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.2 | 4.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 2.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 7.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 7.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 0.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.2 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 3.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 3.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 15.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 6.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 2.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 0.5 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.2 | 3.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 4.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 0.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 1.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.2 | 2.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 16.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 0.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 1.5 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 2.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 0.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.9 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.4 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 1.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 3.7 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 4.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 10.0 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 4.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 2.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 3.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.4 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 3.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.8 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 2.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 5.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 4.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 10.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.7 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.1 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.3 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.1 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 7.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 43.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 4.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 9.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 4.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 7.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 9.3 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0072372 | nonmotile primary cilium(GO:0031513) primary cilium(GO:0072372) |
| 0.0 | 6.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.2 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 5.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 3.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.0 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 1.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0036157 | outer dynein arm(GO:0036157) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.3 | 5.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.2 | 4.8 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 1.2 | 10.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.1 | 5.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.1 | 3.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 1.0 | 9.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.0 | 5.8 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.9 | 5.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.9 | 5.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.9 | 2.7 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.9 | 1.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.9 | 3.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.9 | 14.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.8 | 4.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.8 | 2.5 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.8 | 2.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.8 | 6.3 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.8 | 2.4 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.8 | 2.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.7 | 0.7 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.7 | 2.1 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.7 | 3.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.7 | 2.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.7 | 2.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.7 | 2.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.7 | 2.6 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.7 | 3.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.6 | 3.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.6 | 2.5 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.6 | 3.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.6 | 4.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 1.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.6 | 3.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.6 | 1.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 3.7 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.6 | 4.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.6 | 2.5 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.6 | 3.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.6 | 1.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.6 | 2.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.6 | 2.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.6 | 3.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.5 | 1.6 | GO:0016662 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 0.5 | 3.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.5 | 4.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 1.6 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 6.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 1.6 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.5 | 2.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 2.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.5 | 15.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 1.5 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.5 | 2.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.5 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 1.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.5 | 6.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 13.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.5 | 2.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 0.5 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.5 | 1.5 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.5 | 3.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.5 | 3.9 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.5 | 2.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 1.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.5 | 5.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.5 | 1.9 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.5 | 6.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.5 | 1.4 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.5 | 2.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 2.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 1.8 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.4 | 1.3 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 1.3 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.4 | 1.8 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.4 | 3.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 1.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 1.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.4 | 1.3 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.4 | 8.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 3.0 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.4 | 2.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 16.7 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.4 | 1.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.4 | 1.2 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 2.8 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.4 | 28.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 1.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.4 | 1.2 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.4 | 1.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 2.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 0.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 1.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 7.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 1.5 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.4 | 3.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.4 | 1.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 1.5 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.4 | 1.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.4 | 2.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 1.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 2.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.3 | 1.7 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 1.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 1.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 2.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.3 | 1.0 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.3 | 1.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 10.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 1.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 1.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.3 | 1.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.3 | 1.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 2.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.3 | 2.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 1.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 1.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 2.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 0.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 1.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.3 | 1.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 25.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 2.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 1.6 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 8.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 0.9 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.3 | 2.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 0.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 1.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 3.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 0.9 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 3.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 1.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 1.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.3 | 2.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.3 | 0.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.3 | 1.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.3 | 1.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 1.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 2.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 1.4 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.3 | 1.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 1.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 1.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 0.8 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 6.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 0.8 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 1.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.3 | 2.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 1.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.3 | 4.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.9 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 1.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 1.9 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 5.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 0.8 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 2.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 4.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 1.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 1.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 3.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.2 | 1.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 8.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 7.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 1.0 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.2 | 1.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 21.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 3.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 0.9 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.2 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.9 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.2 | 1.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 0.7 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.2 | 0.7 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.2 | 1.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 2.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.2 | 1.8 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 1.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 0.9 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 0.4 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 1.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 0.7 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 0.7 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 1.5 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.2 | 1.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 2.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 0.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 2.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 1.9 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 7.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.6 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 0.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 1.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 0.6 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.2 | 4.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 3.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 2.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 2.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.2 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 2.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 6.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 1.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 0.8 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 0.6 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.2 | 4.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 0.6 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 5.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.9 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 1.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 11.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 0.5 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.5 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.2 | 0.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 14.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 0.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 2.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 1.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 2.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.2 | 0.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 0.5 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.2 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 0.5 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.2 | 1.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 3.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 2.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 1.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 0.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 1.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.2 | 1.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 6.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.6 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 5.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 3.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.1 | 4.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.7 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.6 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 7.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 4.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.4 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.1 | 1.5 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 4.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0022821 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 10.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 7.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.5 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 1.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 2.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 1.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 1.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.3 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 3.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.7 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 1.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.1 | 1.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.9 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 2.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.6 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.7 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.1 | 1.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 2.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.6 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 2.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.7 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 2.9 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 2.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 2.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.4 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 3.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.5 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.8 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.2 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 3.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.2 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.3 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 2.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 1.8 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.4 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.2 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.2 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 6.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 1.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 2.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 2.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 4.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 64.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 2.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 1.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 6.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 2.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 5.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 7.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.0 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 5.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.8 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 5.2 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0008169 | C-methyltransferase activity(GO:0008169) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.3 | 8.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 10.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 5.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 9.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 4.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 1.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 9.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 5.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 11.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 7.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.6 | 9.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.5 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 17.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 15.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.4 | 10.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 7.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 2.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 3.7 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.3 | 20.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 3.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 4.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 7.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 4.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 4.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 3.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 6.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 7.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 10.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 6.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 3.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 2.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 4.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 3.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 9.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 9.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 6.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.9 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 3.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 0.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 10.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 2.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 10.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.9 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 4.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |