Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zbtb12
Z-value: 1.19
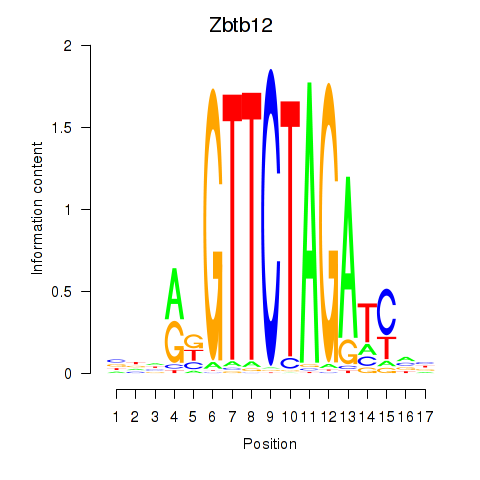
Transcription factors associated with Zbtb12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb12
|
ENSMUSG00000049823.10 | Zbtb12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb12 | mm39_v1_chr17_+_35113490_35113554 | -0.21 | 8.1e-02 | Click! |
Activity profile of Zbtb12 motif
Sorted Z-values of Zbtb12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_36604516 | 17.50 |
ENSMUST00000034620.5
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr15_-_76419131 | 17.18 |
ENSMUST00000230604.2
|
Tmem249
|
transmembrane protein 249 |
| chr5_-_142515792 | 16.75 |
ENSMUST00000099400.3
|
Papolb
|
poly (A) polymerase beta (testis specific) |
| chr11_+_100332709 | 15.41 |
ENSMUST00000001599.4
ENSMUST00000107395.3 |
Klhl10
|
kelch-like 10 |
| chr7_-_126574959 | 12.42 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr7_+_140427711 | 12.31 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr8_+_41128099 | 11.94 |
ENSMUST00000051614.5
|
Adam24
|
a disintegrin and metallopeptidase domain 24 (testase 1) |
| chr9_+_110673565 | 10.81 |
ENSMUST00000176403.8
|
Prss46
|
protease, serine 46 |
| chr18_+_70605722 | 10.69 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr7_+_140427729 | 10.65 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr18_+_70605691 | 10.55 |
ENSMUST00000164223.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr18_+_70605630 | 10.03 |
ENSMUST00000168249.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr5_+_96989853 | 9.71 |
ENSMUST00000196126.2
|
Anxa3
|
annexin A3 |
| chr8_-_44029744 | 9.70 |
ENSMUST00000049577.3
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A (testase 3) |
| chr18_+_70605476 | 9.42 |
ENSMUST00000114959.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr11_+_104983022 | 9.25 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr5_-_121641461 | 9.12 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr14_+_26414422 | 8.99 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr8_+_41246310 | 8.97 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20 |
| chr10_-_75658355 | 8.71 |
ENSMUST00000160211.2
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr4_-_43669141 | 8.70 |
ENSMUST00000056474.7
|
Fam221b
|
family with sequence similarity 221, member B |
| chr7_-_25239229 | 8.61 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr11_+_102775991 | 8.47 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chr3_-_131138541 | 8.38 |
ENSMUST00000090246.5
ENSMUST00000126569.2 |
Sgms2
|
sphingomyelin synthase 2 |
| chr17_+_27248233 | 8.34 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr1_-_161807205 | 8.33 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr18_+_4165832 | 7.99 |
ENSMUST00000025076.10
|
Lyzl1
|
lysozyme-like 1 |
| chr6_+_29398919 | 7.70 |
ENSMUST00000181464.8
ENSMUST00000180829.8 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr8_+_41205245 | 7.53 |
ENSMUST00000096663.5
|
Adam25
|
a disintegrin and metallopeptidase domain 25 (testase 2) |
| chr8_+_71156071 | 7.50 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr6_-_83504756 | 7.30 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr11_-_118021460 | 7.19 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr1_+_69866145 | 7.16 |
ENSMUST00000065425.12
ENSMUST00000113940.4 |
Spag16
|
sperm associated antigen 16 |
| chr15_-_88838605 | 7.07 |
ENSMUST00000109371.8
|
Ttll8
|
tubulin tyrosine ligase-like family, member 8 |
| chr17_-_57181420 | 7.05 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr18_+_4165856 | 6.84 |
ENSMUST00000120837.2
|
Lyzl1
|
lysozyme-like 1 |
| chr11_-_70590923 | 6.72 |
ENSMUST00000108543.4
ENSMUST00000108542.8 ENSMUST00000108541.9 ENSMUST00000126114.9 ENSMUST00000073625.8 |
Inca1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr19_+_6130059 | 6.66 |
ENSMUST00000149347.8
ENSMUST00000143303.2 |
Tmem262
|
transmembrane protein 262 |
| chr8_+_41276027 | 6.47 |
ENSMUST00000066814.7
|
Adam39
|
a disintegrin and metallopeptidase domain 39 |
| chr15_-_76259126 | 6.40 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr6_+_24528143 | 6.39 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr8_-_43981143 | 6.10 |
ENSMUST00000080135.5
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B |
| chrX_+_106836189 | 6.01 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr7_+_99825886 | 5.80 |
ENSMUST00000178946.9
|
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr17_+_79934096 | 5.48 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr15_-_98796373 | 5.14 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr10_+_63293284 | 5.05 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr2_+_148623929 | 4.96 |
ENSMUST00000028933.3
|
Cstdc1
|
cystatin domain containing 1 |
| chr6_-_143892814 | 4.78 |
ENSMUST00000124233.7
ENSMUST00000144289.4 ENSMUST00000111748.8 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr5_+_11234174 | 4.53 |
ENSMUST00000168407.3
|
Gm5861
|
predicted gene 5861 |
| chr6_+_103674695 | 4.21 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr7_+_24476597 | 4.15 |
ENSMUST00000038069.9
ENSMUST00000206847.2 |
Ceacam10
|
carcinoembryonic antigen-related cell adhesion molecule 10 |
| chr11_+_87684299 | 4.11 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr12_-_81615248 | 3.90 |
ENSMUST00000008582.4
|
Adam21
|
a disintegrin and metallopeptidase domain 21 |
| chr11_+_108271990 | 3.45 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr3_-_153610368 | 3.44 |
ENSMUST00000188338.7
|
Msh4
|
mutS homolog 4 |
| chr1_-_75208734 | 3.34 |
ENSMUST00000180101.3
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr3_-_153610528 | 3.28 |
ENSMUST00000190449.2
|
Msh4
|
mutS homolog 4 |
| chr7_+_106413336 | 3.28 |
ENSMUST00000166880.3
ENSMUST00000075414.8 |
Olfr701
|
olfactory receptor 701 |
| chrX_+_71265623 | 3.13 |
ENSMUST00000048675.2
|
Magea4
|
MAGE family member A4 |
| chr4_-_138351313 | 2.84 |
ENSMUST00000030533.12
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr10_-_70428611 | 2.67 |
ENSMUST00000162251.8
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr13_+_104365880 | 2.67 |
ENSMUST00000022227.8
|
Cenpk
|
centromere protein K |
| chr16_-_94657531 | 2.56 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr5_+_30486375 | 2.53 |
ENSMUST00000101448.5
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr15_+_82225380 | 2.37 |
ENSMUST00000050349.3
|
Pheta2
|
PH domain containing endocytic trafficking adaptor 2 |
| chr9_-_122673080 | 2.37 |
ENSMUST00000203176.3
ENSMUST00000203656.3 ENSMUST00000204619.2 |
Gm35549
|
predicted gene, 35549 |
| chr3_-_84063067 | 2.23 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr14_-_79718890 | 2.17 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr2_-_28806639 | 1.97 |
ENSMUST00000113847.3
|
Barhl1
|
BarH like homeobox 1 |
| chr18_-_70605538 | 1.94 |
ENSMUST00000067556.4
|
4930503L19Rik
|
RIKEN cDNA 4930503L19 gene |
| chr7_+_100966289 | 1.87 |
ENSMUST00000163799.9
ENSMUST00000164479.9 |
Stard10
|
START domain containing 10 |
| chr1_+_170472092 | 1.85 |
ENSMUST00000046792.9
|
Olfml2b
|
olfactomedin-like 2B |
| chr13_-_64645606 | 1.71 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr19_-_46561532 | 1.59 |
ENSMUST00000026009.10
ENSMUST00000236255.2 |
Arl3
|
ADP-ribosylation factor-like 3 |
| chr14_+_67470884 | 1.47 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr7_-_48493388 | 1.39 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr4_+_119396913 | 1.37 |
ENSMUST00000137560.8
|
Foxj3
|
forkhead box J3 |
| chr19_+_6919691 | 1.36 |
ENSMUST00000113426.8
ENSMUST00000238184.2 ENSMUST00000113423.10 ENSMUST00000237334.2 ENSMUST00000235240.2 |
Bad
|
BCL2-associated agonist of cell death |
| chr4_-_151142351 | 1.35 |
ENSMUST00000030797.4
|
Vamp3
|
vesicle-associated membrane protein 3 |
| chr17_+_28128307 | 1.30 |
ENSMUST00000233525.2
ENSMUST00000025058.15 ENSMUST00000233866.2 |
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr5_-_35836761 | 1.30 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr2_+_129854256 | 1.29 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr7_+_24596806 | 1.27 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr7_-_35285001 | 1.26 |
ENSMUST00000069912.6
|
Rgs9bp
|
regulator of G-protein signalling 9 binding protein |
| chr17_+_18051266 | 1.25 |
ENSMUST00000172097.9
|
Spaca6
|
sperm acrosome associated 6 |
| chr14_-_70449438 | 1.24 |
ENSMUST00000227929.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr9_+_123902143 | 1.18 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr4_-_119031050 | 1.08 |
ENSMUST00000052715.10
ENSMUST00000179290.8 ENSMUST00000154226.2 |
Zfp691
|
zinc finger protein 691 |
| chr14_+_79718604 | 1.06 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr1_-_155627430 | 0.96 |
ENSMUST00000195275.2
|
Lhx4
|
LIM homeobox protein 4 |
| chr12_-_101924407 | 0.84 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr14_+_26789345 | 0.76 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr11_-_100332619 | 0.74 |
ENSMUST00000107398.8
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr17_+_37253916 | 0.74 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr3_+_87989278 | 0.71 |
ENSMUST00000071812.11
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr14_+_54406112 | 0.68 |
ENSMUST00000103694.3
ENSMUST00000199452.2 |
Traj49
|
T cell receptor alpha joining 49 |
| chr17_-_28128180 | 0.64 |
ENSMUST00000114848.8
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr2_-_150531280 | 0.63 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr6_+_37877413 | 0.61 |
ENSMUST00000120238.2
|
Trim24
|
tripartite motif-containing 24 |
| chr9_+_22322802 | 0.57 |
ENSMUST00000058868.9
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr4_+_43669266 | 0.57 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr9_-_62895197 | 0.53 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chr5_+_92750898 | 0.49 |
ENSMUST00000200941.2
ENSMUST00000050952.4 |
Stbd1
|
starch binding domain 1 |
| chr15_-_60793115 | 0.47 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr3_+_5283577 | 0.45 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr17_-_28779678 | 0.40 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr3_+_5283606 | 0.37 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr4_-_149994176 | 0.37 |
ENSMUST00000105685.2
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr16_-_89368059 | 0.28 |
ENSMUST00000171542.2
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr11_+_69881885 | 0.25 |
ENSMUST00000018711.15
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr12_+_111540920 | 0.24 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr7_+_35285657 | 0.14 |
ENSMUST00000040844.16
ENSMUST00000188906.7 ENSMUST00000186245.7 ENSMUST00000190503.7 |
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr17_+_48607405 | 0.12 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr8_-_70411054 | 0.09 |
ENSMUST00000211960.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr9_+_22322875 | 0.04 |
ENSMUST00000214436.2
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr17_+_83658354 | 0.03 |
ENSMUST00000096766.12
ENSMUST00000049503.10 ENSMUST00000112363.10 ENSMUST00000234460.2 |
Eml4
|
echinoderm microtubule associated protein like 4 |
| chr1_+_38037086 | 0.01 |
ENSMUST00000027252.8
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr7_+_102420428 | 0.00 |
ENSMUST00000213432.2
|
Olfr561
|
olfactory receptor 561 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.4 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 2.6 | 5.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 1.7 | 11.9 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 1.4 | 5.8 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.4 | 4.1 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.2 | 7.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 1.0 | 7.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.8 | 8.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.8 | 14.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.8 | 8.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.7 | 4.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 5.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.5 | 9.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 9.7 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.5 | 1.4 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.5 | 1.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.4 | 3.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 6.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.4 | 1.2 | GO:2000451 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.4 | 7.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 16.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.2 | 1.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 2.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 1.9 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 6.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 0.5 | GO:0061723 | glycophagy(GO:0061723) |
| 0.1 | 7.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 8.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 2.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 4.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 1.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 40.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 58.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 2.6 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 2.2 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 1.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 1.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 1.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 2.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 1.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.6 | 23.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.1 | 6.7 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.7 | 9.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.7 | 7.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.7 | 9.0 | GO:0005858 | axonemal dynein complex(GO:0005858) inner dynein arm(GO:0036156) |
| 0.2 | 4.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 5.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 7.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 6.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 1.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 15.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 8.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 8.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 17.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 8.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 7.3 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.4 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 5.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 5.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 33.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 3.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 1.8 | 7.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 1.4 | 8.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.1 | 9.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.9 | 5.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.8 | 14.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 3.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 16.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 40.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.5 | 6.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.5 | 5.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 1.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.4 | 7.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 6.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 7.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 42.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 2.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 8.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:2001070 | glycogen binding(GO:2001069) starch binding(GO:2001070) |
| 0.1 | 1.4 | GO:0030346 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 5.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 11.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 19.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 15.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 9.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 40.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 8.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 8.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 8.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 5.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 2.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |