Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
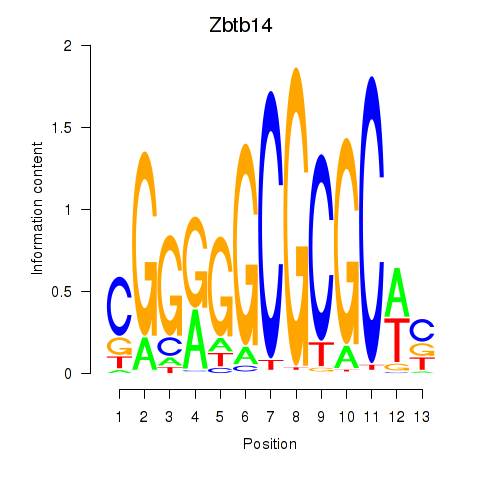
Results for Zbtb14
Z-value: 1.61
Transcription factors associated with Zbtb14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb14
|
ENSMUSG00000049672.16 | Zbtb14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb14 | mm39_v1_chr17_+_69690764_69690835 | 0.47 | 2.9e-05 | Click! |
Activity profile of Zbtb14 motif
Sorted Z-values of Zbtb14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_56703422 | 16.19 |
ENSMUST00000210032.2
|
Lingo1
|
leucine rich repeat and Ig domain containing 1 |
| chr2_+_143388062 | 15.40 |
ENSMUST00000028905.10
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr2_-_117173190 | 14.97 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr1_-_132669490 | 14.92 |
ENSMUST00000094569.11
ENSMUST00000163770.8 ENSMUST00000188307.2 |
Nfasc
|
neurofascin |
| chr9_+_102988940 | 14.57 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr11_-_103844870 | 14.06 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr2_-_117173312 | 13.84 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr17_-_24908874 | 12.38 |
ENSMUST00000007236.5
|
Syngr3
|
synaptogyrin 3 |
| chr15_-_75439013 | 12.03 |
ENSMUST00000156032.2
ENSMUST00000127095.8 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr11_+_7013422 | 10.80 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr7_+_25005510 | 10.78 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chr7_+_123582021 | 10.29 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr2_-_117173428 | 9.97 |
ENSMUST00000102534.11
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chrX_-_156826262 | 9.75 |
ENSMUST00000026750.15
ENSMUST00000112513.2 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr19_+_47167259 | 9.61 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr4_-_117740624 | 9.56 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr15_-_75438457 | 9.44 |
ENSMUST00000163116.8
ENSMUST00000023241.12 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr8_-_100143029 | 9.09 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr15_-_75438660 | 9.00 |
ENSMUST00000065417.15
|
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr16_+_6166982 | 8.97 |
ENSMUST00000056416.9
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr7_-_62862261 | 8.82 |
ENSMUST00000032738.7
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chr1_+_34789662 | 8.75 |
ENSMUST00000159021.9
|
Arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr4_+_42917228 | 8.74 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr2_-_73605684 | 8.68 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr19_-_5148506 | 8.66 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr13_-_49301407 | 8.59 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr4_+_149671012 | 8.55 |
ENSMUST00000039144.7
|
Clstn1
|
calsyntenin 1 |
| chr5_+_37399284 | 8.48 |
ENSMUST00000202434.4
ENSMUST00000114158.9 |
Crmp1
|
collapsin response mediator protein 1 |
| chr4_+_149670889 | 8.48 |
ENSMUST00000105691.8
|
Clstn1
|
calsyntenin 1 |
| chr1_+_182591425 | 8.10 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr15_-_79718462 | 7.94 |
ENSMUST00000148358.2
|
Cbx6
|
chromobox 6 |
| chr7_-_74959010 | 7.94 |
ENSMUST00000165175.8
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr15_+_30173197 | 7.70 |
ENSMUST00000226119.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr9_-_58108988 | 7.68 |
ENSMUST00000163200.3
ENSMUST00000165276.2 ENSMUST00000214647.2 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_-_119438569 | 7.62 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr5_-_146521629 | 7.56 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr9_-_86762467 | 7.49 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr19_+_10366753 | 7.43 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr8_-_110894836 | 7.41 |
ENSMUST00000003754.8
ENSMUST00000212297.2 |
Calb2
|
calbindin 2 |
| chr6_-_113172340 | 7.22 |
ENSMUST00000162280.2
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4 |
| chr8_+_94879235 | 7.16 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chrX_-_63320543 | 7.15 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr4_+_123077515 | 7.13 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr1_+_132808011 | 7.08 |
ENSMUST00000027706.4
|
Lrrn2
|
leucine rich repeat protein 2, neuronal |
| chr17_+_46608333 | 7.05 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr19_+_47003111 | 6.97 |
ENSMUST00000037636.4
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr11_+_104122216 | 6.88 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr10_+_98750268 | 6.81 |
ENSMUST00000219557.2
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr15_+_30172716 | 6.80 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr5_-_139115914 | 6.76 |
ENSMUST00000129851.8
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr6_-_88851579 | 6.75 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr7_-_64806164 | 6.73 |
ENSMUST00000148459.3
ENSMUST00000119118.8 |
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr1_+_63485332 | 6.66 |
ENSMUST00000114103.8
ENSMUST00000114107.3 |
Adam23
|
a disintegrin and metallopeptidase domain 23 |
| chr2_-_104240679 | 6.66 |
ENSMUST00000136156.9
ENSMUST00000141159.9 ENSMUST00000089726.10 |
D430041D05Rik
|
RIKEN cDNA D430041D05 gene |
| chr7_-_105230807 | 6.60 |
ENSMUST00000191011.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr7_-_27095964 | 6.58 |
ENSMUST00000108363.8
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr9_+_87026337 | 6.57 |
ENSMUST00000113149.8
ENSMUST00000049457.14 ENSMUST00000179313.3 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr3_+_31204069 | 6.55 |
ENSMUST00000046174.8
|
Cldn11
|
claudin 11 |
| chr7_+_108610032 | 6.49 |
ENSMUST00000033341.12
|
Tub
|
tubby bipartite transcription factor |
| chr6_-_88852017 | 6.47 |
ENSMUST00000145944.3
|
Podxl2
|
podocalyxin-like 2 |
| chr9_-_21963306 | 6.46 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr11_+_104122399 | 6.39 |
ENSMUST00000132977.8
ENSMUST00000132245.8 ENSMUST00000100347.11 |
Mapt
|
microtubule-associated protein tau |
| chr10_+_20828446 | 6.39 |
ENSMUST00000105525.12
|
Ahi1
|
Abelson helper integration site 1 |
| chr3_-_80710097 | 6.34 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr8_+_122457302 | 6.25 |
ENSMUST00000026357.12
|
Jph3
|
junctophilin 3 |
| chr7_-_105230698 | 6.25 |
ENSMUST00000189378.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr10_+_90665270 | 6.22 |
ENSMUST00000182202.8
ENSMUST00000182966.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_-_79718423 | 6.20 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr2_-_73605387 | 6.14 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr7_-_105230395 | 6.10 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr6_+_22875494 | 6.07 |
ENSMUST00000090568.7
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
| chr19_-_46315543 | 6.04 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr5_-_124939428 | 6.02 |
ENSMUST00000036206.14
|
Ccdc92
|
coiled-coil domain containing 92 |
| chr11_-_66416621 | 5.96 |
ENSMUST00000123454.8
|
Shisa6
|
shisa family member 6 |
| chr3_-_89230190 | 5.93 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr3_-_33197157 | 5.93 |
ENSMUST00000108226.8
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr13_-_58056089 | 5.92 |
ENSMUST00000185502.7
ENSMUST00000186271.7 ENSMUST00000185905.2 ENSMUST00000187852.7 |
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
| chr5_-_38316296 | 5.91 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chr2_-_27032441 | 5.90 |
ENSMUST00000151224.3
|
Fam163b
|
family with sequence similarity 163, member B |
| chr8_+_12965876 | 5.86 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr7_-_105230479 | 5.81 |
ENSMUST00000191601.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr11_-_6015538 | 5.80 |
ENSMUST00000101585.10
ENSMUST00000066431.14 ENSMUST00000109815.9 ENSMUST00000109812.9 ENSMUST00000101586.3 ENSMUST00000093355.12 ENSMUST00000019133.11 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr10_+_90665399 | 5.80 |
ENSMUST00000179694.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_49956441 | 5.78 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr15_+_79975520 | 5.76 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr10_+_90665639 | 5.75 |
ENSMUST00000179337.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr16_-_88087087 | 5.65 |
ENSMUST00000211444.2
ENSMUST00000023652.16 ENSMUST00000072256.13 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr11_+_104122341 | 5.61 |
ENSMUST00000106993.10
|
Mapt
|
microtubule-associated protein tau |
| chr9_+_120132962 | 5.60 |
ENSMUST00000048121.13
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr10_+_58649181 | 5.54 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr18_+_82493284 | 5.52 |
ENSMUST00000047865.14
|
Mbp
|
myelin basic protein |
| chr17_+_46608842 | 5.52 |
ENSMUST00000166617.8
ENSMUST00000170271.2 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr11_+_103061905 | 5.46 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr3_-_125732255 | 5.45 |
ENSMUST00000057944.12
|
Ugt8a
|
UDP galactosyltransferase 8A |
| chr10_+_126914755 | 5.43 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_+_181180183 | 5.41 |
ENSMUST00000161880.8
ENSMUST00000027795.14 |
Cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr8_+_120173458 | 5.39 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr5_+_66833434 | 5.38 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr11_+_17109263 | 5.38 |
ENSMUST00000102880.5
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
| chr4_+_137977714 | 5.37 |
ENSMUST00000105824.8
ENSMUST00000124239.8 ENSMUST00000105823.2 ENSMUST00000105818.8 |
Sh2d5
Kif17
|
SH2 domain containing 5 kinesin family member 17 |
| chr7_-_30750856 | 5.35 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr15_+_34837501 | 5.35 |
ENSMUST00000072868.5
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr2_-_32737238 | 5.34 |
ENSMUST00000050000.16
|
Stxbp1
|
syntaxin binding protein 1 |
| chr12_-_109019507 | 5.28 |
ENSMUST00000185745.2
ENSMUST00000239108.2 |
Begain
|
brain-enriched guanylate kinase-associated |
| chrX_-_72703330 | 5.27 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr1_-_22031718 | 5.24 |
ENSMUST00000029667.13
ENSMUST00000173058.8 ENSMUST00000173404.2 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr11_-_70924288 | 5.20 |
ENSMUST00000238695.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr7_+_131568167 | 5.18 |
ENSMUST00000045840.5
|
Gpr26
|
G protein-coupled receptor 26 |
| chr12_+_74044435 | 5.16 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr5_+_144482693 | 5.13 |
ENSMUST00000071782.8
|
Nptx2
|
neuronal pentraxin 2 |
| chr4_-_41695442 | 5.12 |
ENSMUST00000102961.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr14_+_70768257 | 5.11 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr10_-_80679859 | 5.08 |
ENSMUST00000053986.9
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr2_-_25209107 | 5.06 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr4_-_155445818 | 5.04 |
ENSMUST00000030922.15
|
Prkcz
|
protein kinase C, zeta |
| chr17_+_27904155 | 5.01 |
ENSMUST00000231669.2
ENSMUST00000097360.3 ENSMUST00000231236.2 |
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr2_+_29855572 | 5.01 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr11_-_119937896 | 4.96 |
ENSMUST00000064307.10
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chrX_-_72868544 | 4.96 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr13_-_110416637 | 4.94 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chr9_-_70048766 | 4.94 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr16_-_96971905 | 4.93 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr7_-_84059321 | 4.91 |
ENSMUST00000085077.5
ENSMUST00000207769.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr7_-_30750828 | 4.90 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr8_-_125161061 | 4.90 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr13_+_83672965 | 4.89 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_104241358 | 4.88 |
ENSMUST00000230671.2
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene |
| chrX_-_134968985 | 4.86 |
ENSMUST00000049130.8
|
Bex2
|
brain expressed X-linked 2 |
| chr4_+_129878890 | 4.85 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr10_-_79473675 | 4.84 |
ENSMUST00000020564.7
|
Shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr14_+_70768289 | 4.81 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr9_-_108067552 | 4.79 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr10_+_59942274 | 4.77 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr7_-_27146024 | 4.71 |
ENSMUST00000011895.14
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr17_-_45860580 | 4.71 |
ENSMUST00000180252.3
|
Tmem151b
|
transmembrane protein 151B |
| chr1_+_109911007 | 4.70 |
ENSMUST00000146282.3
|
Cdh7
|
cadherin 7, type 2 |
| chr15_+_100768551 | 4.70 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr1_-_87438027 | 4.69 |
ENSMUST00000027477.15
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr16_+_80997580 | 4.67 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr11_-_119937970 | 4.66 |
ENSMUST00000103020.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr7_-_141649003 | 4.62 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8 |
| chrX_+_100342749 | 4.61 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr11_+_87017878 | 4.59 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr12_+_102094977 | 4.57 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chrX_-_72703652 | 4.56 |
ENSMUST00000114472.8
|
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr18_-_61147272 | 4.55 |
ENSMUST00000025520.10
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chr2_-_32737208 | 4.53 |
ENSMUST00000077458.7
ENSMUST00000208840.2 |
Stxbp1
|
syntaxin binding protein 1 |
| chr7_+_3352019 | 4.52 |
ENSMUST00000100301.11
|
Prkcg
|
protein kinase C, gamma |
| chr4_-_15149051 | 4.51 |
ENSMUST00000041606.14
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr10_-_32765671 | 4.50 |
ENSMUST00000218645.2
|
Nkain2
|
Na+/K+ transporting ATPase interacting 2 |
| chr6_-_57802131 | 4.49 |
ENSMUST00000204878.3
ENSMUST00000145608.7 ENSMUST00000203212.3 ENSMUST00000114297.5 |
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr7_+_99876515 | 4.48 |
ENSMUST00000084935.11
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr15_+_99568208 | 4.46 |
ENSMUST00000023758.9
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr10_-_116309764 | 4.46 |
ENSMUST00000068233.11
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr16_-_9812787 | 4.46 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr11_-_6015736 | 4.44 |
ENSMUST00000002817.12
ENSMUST00000109813.9 ENSMUST00000090443.10 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr11_+_101046708 | 4.44 |
ENSMUST00000043654.10
|
Tubg2
|
tubulin, gamma 2 |
| chr2_+_92430043 | 4.44 |
ENSMUST00000065797.7
|
Chst1
|
carbohydrate sulfotransferase 1 |
| chr8_-_4267260 | 4.42 |
ENSMUST00000168386.9
|
Prr36
|
proline rich 36 |
| chr4_-_49845549 | 4.42 |
ENSMUST00000093859.11
ENSMUST00000076674.4 |
Grin3a
|
glutamate receptor ionotropic, NMDA3A |
| chr4_+_58943574 | 4.38 |
ENSMUST00000107554.2
|
Zkscan16
|
zinc finger with KRAB and SCAN domains 16 |
| chrX_-_58179754 | 4.37 |
ENSMUST00000033473.12
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_+_123077286 | 4.35 |
ENSMUST00000126995.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr2_-_162502994 | 4.31 |
ENSMUST00000109442.8
ENSMUST00000109445.9 ENSMUST00000109443.8 ENSMUST00000109441.2 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr15_-_89726063 | 4.30 |
ENSMUST00000029441.4
|
Syt10
|
synaptotagmin X |
| chr3_+_13536696 | 4.28 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr11_+_29323618 | 4.26 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr9_+_26645141 | 4.23 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr9_+_36744016 | 4.21 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr12_-_109034099 | 4.17 |
ENSMUST00000190647.3
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr13_+_19132375 | 4.15 |
ENSMUST00000239207.2
ENSMUST00000003345.10 ENSMUST00000200466.5 |
Amph
|
amphiphysin |
| chr1_-_38937061 | 4.15 |
ENSMUST00000027249.12
|
Chst10
|
carbohydrate sulfotransferase 10 |
| chr9_+_36743980 | 4.14 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_-_77956499 | 4.14 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr15_+_34838195 | 4.11 |
ENSMUST00000228725.2
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr11_+_104122291 | 4.10 |
ENSMUST00000145227.8
|
Mapt
|
microtubule-associated protein tau |
| chr3_+_58913234 | 4.08 |
ENSMUST00000040846.15
|
Med12l
|
mediator complex subunit 12-like |
| chr14_-_65499835 | 4.06 |
ENSMUST00000131309.3
|
Fzd3
|
frizzled class receptor 3 |
| chr19_+_10366450 | 4.03 |
ENSMUST00000073899.6
|
Syt7
|
synaptotagmin VII |
| chr6_-_119307685 | 4.03 |
ENSMUST00000112756.8
ENSMUST00000068351.14 |
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr2_+_170573727 | 4.02 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
| chr5_-_104059105 | 4.01 |
ENSMUST00000031254.9
|
Klhl8
|
kelch-like 8 |
| chr6_-_122463422 | 4.00 |
ENSMUST00000068242.9
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr1_+_63485002 | 3.99 |
ENSMUST00000087374.10
|
Adam23
|
a disintegrin and metallopeptidase domain 23 |
| chr15_-_94302139 | 3.98 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr2_+_102489558 | 3.97 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_-_112542671 | 3.95 |
ENSMUST00000196256.2
|
Asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr9_+_111140741 | 3.93 |
ENSMUST00000078626.8
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr15_-_78602313 | 3.92 |
ENSMUST00000229441.2
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr5_-_137739863 | 3.90 |
ENSMUST00000061789.14
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr15_+_100768806 | 3.88 |
ENSMUST00000201549.4
ENSMUST00000108908.6 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr14_+_70314727 | 3.87 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr4_-_41695935 | 3.87 |
ENSMUST00000145379.2
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr12_+_117497478 | 3.81 |
ENSMUST00000220781.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr15_+_23036535 | 3.79 |
ENSMUST00000164787.8
|
Cdh18
|
cadherin 18 |
| chr4_-_4138817 | 3.78 |
ENSMUST00000133567.2
|
Penk
|
preproenkephalin |
| chr4_+_42916666 | 3.78 |
ENSMUST00000132173.8
ENSMUST00000107975.8 |
Phf24
|
PHD finger protein 24 |
| chr2_-_31735937 | 3.77 |
ENSMUST00000028188.8
|
Fibcd1
|
fibrinogen C domain containing 1 |
| chr8_+_117983803 | 3.75 |
ENSMUST00000166750.9
|
Cmip
|
c-Maf inducing protein |
| chr7_-_125681577 | 3.74 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0030070 | insulin processing(GO:0030070) |
| 4.2 | 8.5 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 4.1 | 24.8 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 3.8 | 11.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 3.4 | 10.2 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 2.9 | 8.8 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 2.8 | 8.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 2.7 | 8.0 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 2.7 | 40.0 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 2.6 | 7.9 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 2.6 | 26.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.6 | 7.8 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 2.5 | 15.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 2.5 | 10.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 2.4 | 7.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 2.3 | 23.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.2 | 8.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 2.1 | 6.2 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 2.0 | 6.0 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 2.0 | 8.0 | GO:0003360 | brainstem development(GO:0003360) |
| 1.9 | 7.5 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.8 | 10.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.8 | 5.4 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.8 | 5.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.6 | 4.9 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.6 | 17.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.5 | 6.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 1.5 | 6.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 1.4 | 4.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.4 | 9.6 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 1.4 | 4.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 1.4 | 4.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.3 | 4.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.3 | 15.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.2 | 4.9 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.1 | 4.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.1 | 4.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 1.1 | 3.3 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.1 | 6.5 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 1.1 | 8.6 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 1.1 | 5.4 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.0 | 4.1 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 1.0 | 3.0 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 1.0 | 5.9 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 1.0 | 6.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.9 | 30.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.9 | 17.5 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.9 | 7.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.9 | 16.2 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.9 | 3.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.9 | 3.6 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.9 | 7.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.9 | 1.7 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.8 | 2.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.8 | 2.5 | GO:0046959 | habituation(GO:0046959) |
| 0.8 | 1.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.8 | 8.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.8 | 2.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.8 | 2.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.8 | 4.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.7 | 4.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.7 | 5.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.7 | 9.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.7 | 2.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.7 | 2.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.7 | 3.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.7 | 2.9 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.7 | 4.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.7 | 4.9 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.7 | 5.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.7 | 2.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.7 | 5.9 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.7 | 2.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.7 | 4.6 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 6.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.6 | 1.8 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.6 | 8.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.6 | 5.9 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.6 | 4.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.6 | 2.9 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.6 | 1.7 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.6 | 4.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 25.4 | GO:0007616 | long-term memory(GO:0007616) |
| 0.6 | 4.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.6 | 2.8 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.5 | 10.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 1.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.5 | 7.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 15.9 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.5 | 25.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.5 | 5.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 2.0 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.5 | 1.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.5 | 2.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.5 | 3.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.5 | 1.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 1.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 9.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.5 | 1.9 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.5 | 2.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 2.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.5 | 4.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.5 | 1.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.5 | 1.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.5 | 3.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.4 | 13.8 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.4 | 1.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.4 | 1.8 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 0.9 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.4 | 2.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 1.7 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 3.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.7 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 3.8 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 5.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 1.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.4 | 4.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.4 | 2.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.4 | 6.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 6.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.4 | 1.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 2.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 3.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 | 2.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.4 | 2.0 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.4 | 10.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 1.1 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.4 | 1.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 36.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 1.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.4 | 7.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.4 | 2.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.4 | 1.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.4 | 9.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 1.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.3 | 1.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 25.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 2.7 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.3 | 1.7 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.3 | 6.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 1.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 0.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 2.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 9.4 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.3 | 13.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.3 | 3.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 3.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 1.7 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.3 | 1.9 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.3 | 2.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 11.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.3 | 11.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 2.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 | 4.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 4.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 5.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.8 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.3 | 1.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 2.3 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.3 | 3.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.3 | 2.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.3 | 1.7 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.3 | 1.7 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.3 | 3.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 1.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.3 | 10.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 2.9 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.3 | 1.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.3 | 1.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.3 | 1.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.3 | 6.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.3 | 7.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 16.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.3 | 12.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.3 | 1.8 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.3 | 0.8 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 | 1.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 2.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 | 4.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 5.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 1.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 4.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 4.9 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.2 | 2.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 5.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 0.7 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 0.9 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 1.4 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.2 | 0.9 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 7.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 0.7 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 3.9 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 0.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 4.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 8.5 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 1.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 7.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 1.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 0.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 5.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 3.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 1.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 2.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 7.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 0.8 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.2 | 5.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 3.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 13.7 | GO:0008306 | associative learning(GO:0008306) |
| 0.2 | 1.4 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 1.7 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 2.9 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 8.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 2.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.7 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 4.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.2 | 1.9 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 1.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 5.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 1.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 5.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.6 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 3.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 2.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 3.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 2.5 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.1 | 1.6 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.6 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.1 | 1.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 3.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 3.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 12.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.5 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 4.3 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 0.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.5 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 1.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 2.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.8 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 2.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.5 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 3.6 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 1.6 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 0.4 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 1.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 1.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.6 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 0.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.8 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:0021815 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.1 | 1.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.7 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 6.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 1.8 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.3 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 1.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.9 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.1 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.8 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 1.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.5 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.1 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.7 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.1 | 2.7 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 5.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.1 | 1.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.3 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.1 | 1.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 10.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 2.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.1 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 3.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 2.1 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.1 | 3.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 2.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 2.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 2.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.0 | GO:0090308 | maintenance of DNA methylation(GO:0010216) regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 5.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 6.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 7.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.6 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.0 | 1.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 1.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.9 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 1.0 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.4 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.0 | 0.6 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.7 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 9.4 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 1.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.7 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.5 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0019348 | polyprenol catabolic process(GO:0016095) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process(GO:0009148) |
| 0.0 | 0.2 | GO:0021707 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0097402 | optic vesicle morphogenesis(GO:0003404) neuroblast migration(GO:0097402) |
| 0.0 | 0.1 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.5 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 | 0.2 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.8 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.5 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 3.3 | 36.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 2.9 | 14.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.6 | 23.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.1 | 6.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 1.8 | 9.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.7 | 6.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.6 | 11.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.6 | 8.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.6 | 4.8 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.5 | 6.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.4 | 11.3 | GO:0008091 | spectrin(GO:0008091) |
| 1.4 | 12.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.3 | 5.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.2 | 3.6 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 1.0 | 21.9 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 1.0 | 3.0 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.0 | 2.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.9 | 10.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.9 | 7.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.8 | 20.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 3.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.7 | 5.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.7 | 2.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.7 | 5.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.7 | 5.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.6 | 9.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 16.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 9.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 8.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 7.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.6 | 1.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 7.4 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.5 | 4.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.5 | 20.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 19.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 7.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 5.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 1.9 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.5 | 2.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 4.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.4 | 2.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 1.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 6.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 4.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 43.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 6.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 3.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.3 | 7.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 5.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 1.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 2.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 58.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 28.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 8.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.3 | 4.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 13.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 2.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 1.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 15.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 5.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 2.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 62.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 2.4 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.2 | 1.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 5.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 1.4 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.2 | 21.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 25.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 7.8 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.2 | 2.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 44.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 3.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 3.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 5.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 4.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 4.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 2.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 5.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 13.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 5.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 18.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.8 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 10.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 3.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.3 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 11.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 9.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 30.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 13.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 2.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 3.2 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.9 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 5.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 2.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 36.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0044440 | endosomal part(GO:0044440) |
| 0.0 | 1.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 4.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 23.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 4.0 | 16.0 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 3.6 | 36.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 3.2 | 38.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 3.2 | 9.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 2.3 | 13.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.2 | 6.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 2.1 | 8.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 2.1 | 14.8 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.5 | 1.5 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.5 | 4.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.5 | 5.8 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 1.4 | 4.1 | GO:0016232 | HNK-1 sulfotransferase activity(GO:0016232) |
| 1.4 | 14.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.3 | 8.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.3 | 5.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.2 | 7.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.2 | 16.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.2 | 7.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.1 | 10.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.1 | 7.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.1 | 6.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.1 | 7.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.0 | 4.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.0 | 3.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.0 | 19.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 1.0 | 5.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.8 | 6.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.8 | 5.9 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.8 | 2.4 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.8 | 2.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.8 | 25.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.8 | 22.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.7 | 4.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.7 | 3.0 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.7 | 2.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.7 | 6.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.7 | 10.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.7 | 2.9 | GO:0047238 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.7 | 20.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.7 | 2.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.7 | 2.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.7 | 4.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 5.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.7 | 9.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 5.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.6 | 2.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.6 | 10.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 2.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.6 | 4.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 42.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.6 | 15.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.6 | 3.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.6 | 1.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.5 | 4.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 2.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.5 | 3.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 4.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.5 | 2.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.5 | 6.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 1.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.5 | 2.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.5 | 5.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 1.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 2.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.5 | 8.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 6.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 21.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.5 | 4.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.4 | 1.8 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.3 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.4 | 2.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.4 | 6.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 4.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 2.5 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 5.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 1.2 | GO:0071820 | N-box binding(GO:0071820) |
| 0.4 | 8.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 7.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 3.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 4.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 3.3 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.4 | 3.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 4.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 7.6 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.3 | 1.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 7.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 1.0 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 5.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 5.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 1.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 1.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.3 | 4.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 13.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 3.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 3.8 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 22.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 11.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 23.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 8.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 1.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 2.0 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.3 | 2.0 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 10.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 4.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 8.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 1.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 6.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 7.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 11.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 2.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 0.8 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.3 | 1.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 1.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 1.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 5.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 1.0 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 0.9 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 4.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 0.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 2.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 15.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 12.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 2.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.2 | 0.8 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.2 | 1.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 3.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 2.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.0 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 1.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 1.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.8 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 11.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 1.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 4.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 3.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 3.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 1.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 0.7 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 7.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 3.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 13.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.2 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 5.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 2.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 8.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 11.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 2.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.7 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 6.1 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 2.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.5 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.1 | 11.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 13.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 5.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 7.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 2.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 4.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 2.9 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 2.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 3.2 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 1.0 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 4.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.7 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 12.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.4 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 3.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 16.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 56.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 2.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.2 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 2.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.6 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 3.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 23.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.9 | 52.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.9 | 21.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 16.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 14.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 22.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 9.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 8.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 11.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 6.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 22.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 4.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 18.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 5.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 3.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 6.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 6.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 4.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 13.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 17.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 4.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 11.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 45.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 1.1 | 25.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.0 | 16.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.9 | 22.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.9 | 12.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.8 | 15.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.7 | 26.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 8.8 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.6 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 9.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.6 | 28.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 9.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 17.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 11.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 16.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.4 | 16.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 6.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 16.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 7.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.3 | 4.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 3.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 4.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 3.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 14.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 4.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.2 | 15.7 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.2 | 3.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.2 | 4.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 1.9 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.2 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 9.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 5.7 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 19.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 11.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 14.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 7.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 9.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 3.5 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.8 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |