Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zbtb16
Z-value: 1.08
Transcription factors associated with Zbtb16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb16
|
ENSMUSG00000066687.6 | Zbtb16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb16 | mm39_v1_chr9_-_48747474_48747563 | -0.49 | 1.4e-05 | Click! |
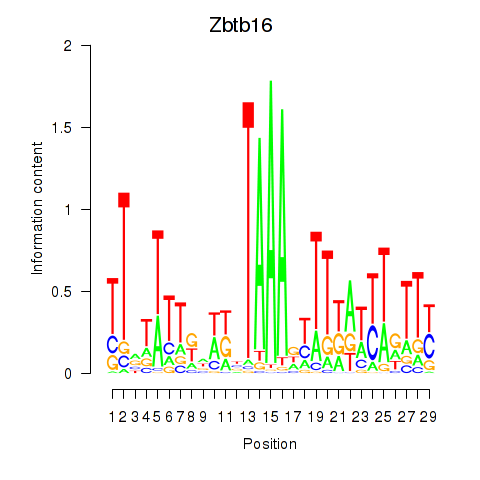
Activity profile of Zbtb16 motif
Sorted Z-values of Zbtb16 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb16
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_169973076 | 7.62 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr1_-_139487951 | 7.02 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr5_-_139799953 | 5.56 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr16_-_97315973 | 5.47 |
ENSMUST00000232018.2
|
Fam3b
|
family with sequence similarity 3, member B |
| chr7_-_12731594 | 5.33 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr5_+_115604321 | 5.32 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr5_+_90666791 | 5.27 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr5_+_115061293 | 5.06 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr16_-_97316013 | 4.99 |
ENSMUST00000231641.2
|
Fam3b
|
family with sequence similarity 3, member B |
| chr8_-_41668182 | 4.73 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chr5_-_77359039 | 4.62 |
ENSMUST00000121825.2
|
Spink2
|
serine peptidase inhibitor, Kazal type 2 |
| chr11_-_5900019 | 4.61 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr4_-_73709231 | 4.60 |
ENSMUST00000222414.2
|
Rasef
|
RAS and EF hand domain containing |
| chr6_+_137731526 | 4.48 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr12_-_103423472 | 4.37 |
ENSMUST00000044687.7
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr10_+_62860291 | 4.37 |
ENSMUST00000020262.5
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr17_+_28749780 | 4.35 |
ENSMUST00000233923.2
|
Armc12
|
armadillo repeat containing 12 |
| chrX_-_94521712 | 4.35 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr9_-_71075939 | 4.31 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr7_-_121785385 | 4.29 |
ENSMUST00000033153.9
|
Ern2
|
endoplasmic reticulum (ER) to nucleus signalling 2 |
| chr17_+_28749808 | 4.26 |
ENSMUST00000233837.2
ENSMUST00000025060.4 |
Armc12
|
armadillo repeat containing 12 |
| chr11_+_87685032 | 4.13 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr2_+_68490186 | 4.11 |
ENSMUST00000055930.6
|
4932414N04Rik
|
RIKEN cDNA 4932414N04 gene |
| chr2_-_25517945 | 4.02 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr15_+_54274151 | 3.84 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr5_-_139799780 | 3.79 |
ENSMUST00000146780.3
|
Tmem184a
|
transmembrane protein 184a |
| chr4_-_117035922 | 3.67 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr1_+_74430575 | 3.64 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr3_+_146276147 | 3.64 |
ENSMUST00000199489.5
|
Uox
|
urate oxidase |
| chr1_+_157334347 | 3.50 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334298 | 3.39 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr6_+_40619913 | 3.33 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr5_+_119809076 | 3.21 |
ENSMUST00000018748.9
|
Tbx3
|
T-box 3 |
| chr7_+_79896121 | 3.18 |
ENSMUST00000058266.9
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr17_+_24072493 | 3.13 |
ENSMUST00000061725.8
|
Prss32
|
protease, serine 32 |
| chr4_-_107975723 | 3.12 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr2_-_6134926 | 3.08 |
ENSMUST00000126551.2
ENSMUST00000054254.12 ENSMUST00000114942.9 |
Proser2
|
proline and serine rich 2 |
| chr4_-_61592331 | 2.90 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr7_-_14180496 | 2.89 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr1_+_171246593 | 2.86 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr2_-_125993887 | 2.82 |
ENSMUST00000110448.3
ENSMUST00000110446.9 |
Fam227b
|
family with sequence similarity 227, member B |
| chr1_+_127657142 | 2.82 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr9_-_118986123 | 2.80 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr4_-_107975701 | 2.78 |
ENSMUST00000149106.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr14_-_66071412 | 2.77 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr13_+_55300453 | 2.76 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr4_-_133954669 | 2.73 |
ENSMUST00000105878.3
ENSMUST00000055892.10 ENSMUST00000169381.8 |
Catsper4
|
cation channel, sperm associated 4 |
| chr11_-_83469446 | 2.71 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr1_+_31261889 | 2.69 |
ENSMUST00000027230.3
|
Dnaaf6
|
dynein axonemal assembly factor 6 |
| chr15_+_100178718 | 2.66 |
ENSMUST00000096200.6
|
Tmprss12
|
transmembrane (C-terminal) protease, serine 12 |
| chr13_+_93810911 | 2.58 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr1_-_139786421 | 2.56 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr9_+_118931532 | 2.55 |
ENSMUST00000165231.8
ENSMUST00000140326.8 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr17_-_36343573 | 2.55 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr2_+_80122811 | 2.53 |
ENSMUST00000057072.6
|
Prdx6b
|
peroxiredoxin 6B |
| chrY_-_84576176 | 2.52 |
ENSMUST00000189463.2
|
Gm21394
|
predicted gene, 21394 |
| chr6_-_131224305 | 2.50 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr2_-_58050494 | 2.50 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr7_-_115445315 | 2.49 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr15_-_36164963 | 2.47 |
ENSMUST00000227793.2
|
Fbxo43
|
F-box protein 43 |
| chr9_+_50528813 | 2.47 |
ENSMUST00000141366.8
|
Pih1d2
|
PIH1 domain containing 2 |
| chr3_-_87985602 | 2.45 |
ENSMUST00000050258.9
|
Ttc24
|
tetratricopeptide repeat domain 24 |
| chr9_-_107556823 | 2.45 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chrY_-_78849325 | 2.45 |
ENSMUST00000190349.2
|
Gm20806
|
predicted gene, 20806 |
| chr2_-_111059901 | 2.45 |
ENSMUST00000028577.3
|
A26c3
|
ANKRD26-like family C, member 3 |
| chrX_+_64091244 | 2.39 |
ENSMUST00000033524.3
|
Ctag2
|
cancer/testis antigen 2 |
| chr15_+_85620308 | 2.38 |
ENSMUST00000057979.6
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr4_+_109272828 | 2.37 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chrY_-_70284911 | 2.37 |
ENSMUST00000187418.2
|
Gm21118
|
predicted gene, 21118 |
| chrX_+_163289318 | 2.32 |
ENSMUST00000033756.3
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr13_-_93810808 | 2.30 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr15_-_36165017 | 2.29 |
ENSMUST00000058643.4
|
Fbxo43
|
F-box protein 43 |
| chrY_-_66753392 | 2.28 |
ENSMUST00000188011.2
|
Gm20852
|
predicted gene, 20852 |
| chr7_-_109330915 | 2.28 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chr12_+_35034747 | 2.27 |
ENSMUST00000134550.3
|
Prps1l1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr19_-_5452521 | 2.26 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr3_-_157630690 | 2.25 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chrY_-_57542272 | 2.24 |
ENSMUST00000178181.3
|
Gm21943
|
predicted gene, 21943 |
| chr10_-_23112973 | 2.21 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chrY_+_21164570 | 2.21 |
ENSMUST00000181205.2
|
Gm20909
|
predicted gene, 20909 |
| chrY_-_79162391 | 2.20 |
ENSMUST00000188706.2
|
Gm20917
|
predicted gene, 20917 |
| chr19_-_30152814 | 2.20 |
ENSMUST00000025778.9
|
Gldc
|
glycine decarboxylase |
| chr11_-_32477551 | 2.19 |
ENSMUST00000054327.3
|
Efcab9
|
EF-hand calcium binding domain 9 |
| chr2_-_72817060 | 2.16 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chr8_-_73197616 | 2.15 |
ENSMUST00000019876.12
|
Calr3
|
calreticulin 3 |
| chrY_-_67059341 | 2.12 |
ENSMUST00000190565.2
|
Ssty2
|
spermiogenesis specific transcript on the Y 2 |
| chrY_-_53414870 | 2.10 |
ENSMUST00000187856.2
|
Gm20747
|
predicted gene, 20747 |
| chr4_-_119241002 | 2.09 |
ENSMUST00000238721.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr7_-_45173193 | 2.07 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_-_140597837 | 2.03 |
ENSMUST00000209328.2
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr11_+_101877876 | 2.02 |
ENSMUST00000010985.8
|
Cfap97d1
|
CFAP97 domain containing 1 |
| chr3_-_146321341 | 2.02 |
ENSMUST00000200633.2
|
Dnase2b
|
deoxyribonuclease II beta |
| chr7_-_115445352 | 1.99 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chrY_-_8835169 | 1.95 |
ENSMUST00000115496.4
|
Gm20815
|
predicted gene, 20815 |
| chr4_-_140867038 | 1.95 |
ENSMUST00000148204.8
ENSMUST00000102487.4 |
Szrd1
|
SUZ RNA binding domain containing 1 |
| chr4_-_19570073 | 1.94 |
ENSMUST00000029885.5
|
Cpne3
|
copine III |
| chr4_+_136337742 | 1.91 |
ENSMUST00000046647.3
|
Tex46
|
testis expressed 46 |
| chr5_-_123620632 | 1.91 |
ENSMUST00000198901.2
|
Il31
|
interleukin 31 |
| chr5_-_5713264 | 1.91 |
ENSMUST00000148193.2
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr15_+_79555272 | 1.91 |
ENSMUST00000127292.2
|
Tomm22
|
translocase of outer mitochondrial membrane 22 |
| chrY_-_8071507 | 1.90 |
ENSMUST00000178149.3
|
Gm20825
|
predicted gene, 20825 |
| chr10_+_68987257 | 1.90 |
ENSMUST00000167286.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr4_-_119241024 | 1.90 |
ENSMUST00000127149.8
ENSMUST00000152879.9 ENSMUST00000238673.2 ENSMUST00000238485.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr5_-_18093739 | 1.88 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chrX_-_9335525 | 1.87 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr10_+_22236451 | 1.87 |
ENSMUST00000182677.8
|
Raet1d
|
retinoic acid early transcript delta |
| chrY_-_85542177 | 1.86 |
ENSMUST00000181549.2
|
Gm20854
|
predicted gene, 20854 |
| chr17_+_49735386 | 1.84 |
ENSMUST00000165390.9
ENSMUST00000024797.16 |
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr12_-_36303394 | 1.84 |
ENSMUST00000221155.2
|
Lrrc72
|
leucine rich repeat containing 72 |
| chr18_+_77861656 | 1.82 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr17_+_49735413 | 1.81 |
ENSMUST00000173033.8
|
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr12_+_65272495 | 1.81 |
ENSMUST00000221980.2
|
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr12_-_75678092 | 1.80 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr1_-_82746169 | 1.80 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr8_+_34007333 | 1.79 |
ENSMUST00000124496.8
|
Tex15
|
testis expressed gene 15 |
| chr2_-_168607166 | 1.76 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr19_-_8382424 | 1.74 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr4_+_116544509 | 1.74 |
ENSMUST00000030454.6
|
Prdx1
|
peroxiredoxin 1 |
| chr17_-_29162794 | 1.73 |
ENSMUST00000232977.2
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr9_-_37623573 | 1.71 |
ENSMUST00000104875.2
|
Olfr160
|
olfactory receptor 160 |
| chrY_-_9347184 | 1.70 |
ENSMUST00000188908.2
|
Gm21812
|
predicted gene, 21812 |
| chr14_+_43156329 | 1.69 |
ENSMUST00000228117.2
|
Gm9732
|
predicted gene 9732 |
| chr2_+_11710101 | 1.69 |
ENSMUST00000138349.8
ENSMUST00000135341.8 ENSMUST00000128156.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr11_-_5657658 | 1.69 |
ENSMUST00000154330.2
|
Mrps24
|
mitochondrial ribosomal protein S24 |
| chr5_-_106606032 | 1.67 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr11_+_11439266 | 1.66 |
ENSMUST00000109678.3
|
Spata48
|
spermatogenesis associated 48 |
| chr1_+_107439145 | 1.64 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr10_-_13053733 | 1.63 |
ENSMUST00000019954.6
|
Zc2hc1b
|
zinc finger, C2HC-type containing 1B |
| chr1_-_93659622 | 1.62 |
ENSMUST00000189728.7
|
Thap4
|
THAP domain containing 4 |
| chr8_+_85635189 | 1.61 |
ENSMUST00000003910.13
ENSMUST00000109744.8 |
Dnase2a
|
deoxyribonuclease II alpha |
| chr12_+_65272287 | 1.60 |
ENSMUST00000046331.5
ENSMUST00000221658.2 |
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr5_-_129781323 | 1.59 |
ENSMUST00000042266.13
|
Septin14
|
septin 14 |
| chr9_+_50528608 | 1.59 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr1_+_170136372 | 1.59 |
ENSMUST00000056991.6
|
Spata46
|
spermatogenesis associated 46 |
| chrY_-_1286623 | 1.57 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr4_+_129181407 | 1.56 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr2_-_37007795 | 1.55 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr5_-_121045568 | 1.55 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr8_+_34006758 | 1.53 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chrX_-_6907858 | 1.53 |
ENSMUST00000115752.8
|
Ccnb3
|
cyclin B3 |
| chr9_+_96140781 | 1.53 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr1_+_58841808 | 1.52 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr12_-_44257109 | 1.52 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr18_-_35795175 | 1.51 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr1_+_58841650 | 1.50 |
ENSMUST00000165549.8
|
Casp8
|
caspase 8 |
| chr6_-_70094604 | 1.50 |
ENSMUST00000103378.3
|
Igkv8-30
|
immunoglobulin kappa chain variable 8-30 |
| chr13_+_30320446 | 1.49 |
ENSMUST00000047311.16
|
Mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr1_-_31261678 | 1.49 |
ENSMUST00000187892.8
ENSMUST00000233331.2 |
4931428L18Rik
|
RIKEN cDNA 4931428L18 gene |
| chrX_-_136022832 | 1.47 |
ENSMUST00000155283.2
|
Esx1
|
extraembryonic, spermatogenesis, homeobox 1 |
| chrX_+_85087792 | 1.46 |
ENSMUST00000047945.2
|
Samt3
|
spermatogenesis associated multipass transmembrane protein 3 |
| chr6_-_142647985 | 1.45 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_+_146599377 | 1.45 |
ENSMUST00000140089.7
ENSMUST00000179175.3 |
Zfp981
|
zinc finger protein 981 |
| chr17_-_14279793 | 1.45 |
ENSMUST00000186636.3
|
Gm7358
|
predicted gene 7358 |
| chr14_+_44523037 | 1.45 |
ENSMUST00000164663.8
ENSMUST00000169601.3 |
Gm8220
|
predicted gene 8220 |
| chr7_+_45267024 | 1.42 |
ENSMUST00000008605.6
ENSMUST00000209379.2 |
Fut1
|
fucosyltransferase 1 |
| chr11_-_74243447 | 1.42 |
ENSMUST00000141134.2
ENSMUST00000214769.2 |
Olfr411
|
olfactory receptor 411 |
| chr4_+_134847949 | 1.41 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr19_+_39102342 | 1.40 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr11_+_114559350 | 1.39 |
ENSMUST00000106602.10
ENSMUST00000077915.10 ENSMUST00000106599.8 ENSMUST00000082092.5 |
Rpl38
|
ribosomal protein L38 |
| chr6_-_142647944 | 1.39 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chrX_-_52759798 | 1.38 |
ENSMUST00000067940.3
|
1700013H16Rik
|
RIKEN cDNA 1700013H16 gene |
| chr18_-_67582191 | 1.36 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr6_-_42437951 | 1.33 |
ENSMUST00000090156.2
|
Olfr458
|
olfactory receptor 458 |
| chr6_-_124613044 | 1.33 |
ENSMUST00000068797.3
ENSMUST00000218020.2 |
C1s2
|
complement component 1, s subcomponent 2 |
| chr5_-_105130522 | 1.33 |
ENSMUST00000031239.13
|
Abcg3
|
ATP binding cassette subfamily G member 3 |
| chr16_+_36097313 | 1.32 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr13_+_55875158 | 1.27 |
ENSMUST00000021958.6
ENSMUST00000124968.8 |
Pcbd2
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr7_+_4795873 | 1.27 |
ENSMUST00000032597.12
ENSMUST00000078432.5 |
Rpl28
|
ribosomal protein L28 |
| chr7_+_102834010 | 1.26 |
ENSMUST00000106893.3
|
Olfr592
|
olfactory receptor 592 |
| chr17_+_38234397 | 1.26 |
ENSMUST00000080231.5
|
Olfr128
|
olfactory receptor 128 |
| chr14_+_44602814 | 1.26 |
ENSMUST00000168161.2
|
Gm8229
|
predicted gene 8229 |
| chr13_+_52737473 | 1.26 |
ENSMUST00000118756.8
|
Syk
|
spleen tyrosine kinase |
| chr7_-_14226851 | 1.25 |
ENSMUST00000108524.4
ENSMUST00000211740.2 ENSMUST00000209744.2 |
Sult2a7
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 7 |
| chr9_-_44473146 | 1.25 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr14_+_44435118 | 1.24 |
ENSMUST00000166350.2
|
Gm8212
|
predicted gene 8212 |
| chrY_+_50093855 | 1.24 |
ENSMUST00000187033.3
|
Gm28870
|
predicted gene 28870 |
| chr3_+_84081411 | 1.23 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr6_+_116483477 | 1.23 |
ENSMUST00000075756.3
|
Olfr212
|
olfactory receptor 212 |
| chr16_-_92494203 | 1.23 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr4_-_116678904 | 1.23 |
ENSMUST00000055436.5
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr15_-_5093222 | 1.21 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr15_-_103785705 | 1.20 |
ENSMUST00000078842.11
ENSMUST00000226484.2 |
Mucl1
|
mucin-like 1 |
| chr16_+_17149235 | 1.20 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr9_+_96141317 | 1.20 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr17_-_41191414 | 1.19 |
ENSMUST00000068258.3
ENSMUST00000233122.2 |
9130008F23Rik
|
RIKEN cDNA 9130008F23 gene |
| chr11_-_99328969 | 1.19 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr9_+_96141299 | 1.18 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr11_+_68936457 | 1.18 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr9_+_44990447 | 1.18 |
ENSMUST00000050020.8
|
Jaml
|
junction adhesion molecule like |
| chrX_-_4194587 | 1.17 |
ENSMUST00000179325.2
|
Btbd35f20
|
BTB domain containing 35, family member 20 |
| chr18_-_3299452 | 1.17 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr13_-_21823691 | 1.16 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr10_-_127587576 | 1.13 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr1_-_171362852 | 1.13 |
ENSMUST00000043094.13
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr9_+_96140750 | 1.12 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr19_+_4008645 | 1.09 |
ENSMUST00000179433.8
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr6_+_68279392 | 1.08 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr11_-_106670309 | 1.08 |
ENSMUST00000021060.6
|
Polg2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr17_+_36190662 | 1.07 |
ENSMUST00000025292.15
|
Dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr5_-_104169785 | 1.07 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.6 | 4.8 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 1.6 | 9.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 1.5 | 5.9 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 1.4 | 4.1 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.3 | 5.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.1 | 4.3 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.1 | 3.2 | GO:0003167 | atrioventricular bundle cell differentiation(GO:0003167) |
| 0.9 | 4.6 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.9 | 2.8 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.8 | 2.5 | GO:2000566 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.8 | 7.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.7 | 4.5 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.7 | 2.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.7 | 2.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.7 | 6.9 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.6 | 4.5 | GO:0071265 | L-methionine biosynthetic process(GO:0071265) |
| 0.6 | 3.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 2.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.5 | 4.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.5 | 2.4 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.5 | 2.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.5 | 3.7 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.4 | 4.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 5.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 2.5 | GO:0051342 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.4 | 1.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.4 | 1.9 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.3 | 1.0 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.3 | 1.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.3 | 1.3 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.3 | 1.2 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 0.9 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.3 | 2.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.2 | 1.9 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.2 | 4.0 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 3.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 | 1.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.2 | 2.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 4.7 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 3.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 0.7 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 0.7 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.2 | 0.7 | GO:1901491 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of lymphangiogenesis(GO:1901491) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.4 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.2 | 0.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 1.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.4 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.1 | 2.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.6 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 1.4 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 1.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 2.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 3.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.3 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 1.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.7 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0009216 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.9 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 5.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.8 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 1.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.8 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 1.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 1.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.0 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 2.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.3 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 2.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.7 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.1 | 2.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 2.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 6.2 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 2.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 3.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 3.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 1.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 3.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.2 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) peptidyl-glutamine methylation(GO:0018364) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 1.7 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 1.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 10.5 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.9 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 1.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.7 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 2.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.5 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.7 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 4.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 1.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.0 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 1.6 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.7 | 4.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.6 | 10.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 3.0 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.5 | 1.4 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.5 | 5.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 1.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 2.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 0.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 4.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 4.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 1.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 2.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 1.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 2.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 2.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 9.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.8 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 3.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 8.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 7.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 5.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 15.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 2.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 6.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 3.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 4.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 4.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.7 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.5 | 5.9 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.9 | 2.8 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.9 | 4.3 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.8 | 3.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.7 | 3.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 2.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.6 | 3.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.6 | 6.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.5 | 2.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 4.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 1.4 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.5 | 4.6 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.5 | 1.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 1.3 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.4 | 1.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 1.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 3.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 5.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 2.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 3.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 1.4 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.3 | 2.8 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 2.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 2.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 1.6 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 0.7 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.2 | 4.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 0.8 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 2.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 3.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 2.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 2.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 3.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 3.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 4.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 1.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 1.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 1.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.4 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 1.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 2.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 4.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 3.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 15.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 7.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 6.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0023023 | MHC protein complex binding(GO:0023023) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 2.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 5.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 3.0 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 5.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 5.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 6.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 11.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 6.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 4.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 4.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 5.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 2.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 3.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |