Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
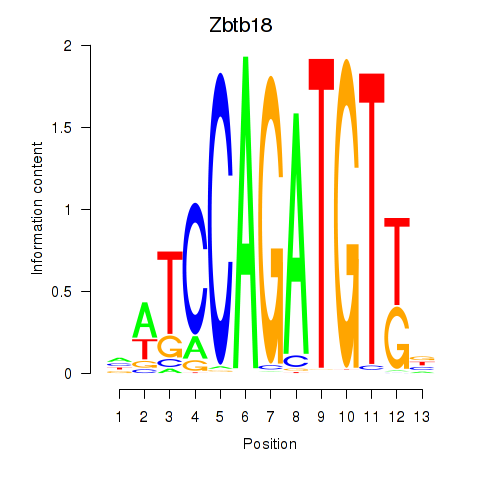
Results for Zbtb18
Z-value: 1.62
Transcription factors associated with Zbtb18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb18
|
ENSMUSG00000063659.12 | Zbtb18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb18 | mm39_v1_chr1_+_177273226_177273387 | -0.65 | 4.6e-10 | Click! |
Activity profile of Zbtb18 motif
Sorted Z-values of Zbtb18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_30608433 | 24.76 |
ENSMUST00000120269.11
ENSMUST00000078490.14 ENSMUST00000006703.15 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr14_+_30608478 | 22.63 |
ENSMUST00000168782.4
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr10_+_62897353 | 19.06 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr2_+_43445333 | 19.02 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr6_+_37507108 | 17.07 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr12_-_103956176 | 14.40 |
ENSMUST00000151709.3
ENSMUST00000176246.3 ENSMUST00000074693.13 ENSMUST00000120251.9 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr1_+_160806241 | 13.90 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr2_+_43445359 | 13.69 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr9_-_103097022 | 13.06 |
ENSMUST00000168142.8
|
Trf
|
transferrin |
| chr4_-_115353326 | 12.82 |
ENSMUST00000030487.3
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr7_-_14180496 | 12.73 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr1_-_139786421 | 12.25 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr7_-_14180576 | 12.15 |
ENSMUST00000125941.3
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr17_-_12894716 | 11.81 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr4_-_49506538 | 11.63 |
ENSMUST00000043056.9
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chr11_+_120421496 | 11.56 |
ENSMUST00000026119.8
|
Gcgr
|
glucagon receptor |
| chr17_-_84154196 | 11.21 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr14_+_40826970 | 10.85 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr1_-_139708906 | 10.79 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr1_+_160806194 | 10.04 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr2_-_25390625 | 9.55 |
ENSMUST00000040042.11
|
C8g
|
complement component 8, gamma polypeptide |
| chr6_-_142418801 | 9.02 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr5_-_145521533 | 8.87 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr1_-_162694076 | 8.76 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr11_+_98938137 | 8.67 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr14_+_40827108 | 8.64 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr4_+_115375461 | 8.45 |
ENSMUST00000058785.10
ENSMUST00000094886.4 |
Cyp4a10
|
cytochrome P450, family 4, subfamily a, polypeptide 10 |
| chr14_-_31362909 | 8.41 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr15_-_96918203 | 8.33 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr14_+_40873399 | 8.26 |
ENSMUST00000225792.2
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr14_+_40827317 | 8.15 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr14_-_31362835 | 8.12 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr14_+_36776775 | 7.82 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr6_+_72332423 | 7.39 |
ENSMUST00000069695.9
ENSMUST00000132243.3 |
Tmem150a
|
transmembrane protein 150A |
| chr5_-_145656934 | 7.31 |
ENSMUST00000094111.6
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr8_+_13076024 | 7.16 |
ENSMUST00000033820.4
|
F7
|
coagulation factor VII |
| chr5_-_145406533 | 7.13 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr17_-_84990360 | 7.10 |
ENSMUST00000066175.10
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr8_-_25066313 | 7.00 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr6_+_72332449 | 6.92 |
ENSMUST00000206064.2
|
Tmem150a
|
transmembrane protein 150A |
| chr10_-_25412010 | 6.91 |
ENSMUST00000179685.3
|
Smlr1
|
small leucine-rich protein 1 |
| chr9_+_65537834 | 6.65 |
ENSMUST00000055844.10
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr4_+_115458172 | 6.48 |
ENSMUST00000084342.6
|
Cyp4a32
|
cytochrome P450, family 4, subfamily a, polypeptide 32 |
| chr7_-_19432933 | 6.47 |
ENSMUST00000174355.8
ENSMUST00000172983.8 ENSMUST00000174710.2 ENSMUST00000003066.16 ENSMUST00000174064.9 |
Apoe
|
apolipoprotein E |
| chr13_+_30520416 | 6.33 |
ENSMUST00000222503.2
ENSMUST00000222370.2 ENSMUST00000066412.8 ENSMUST00000223201.2 |
Agtr1a
|
angiotensin II receptor, type 1a |
| chr1_-_150341911 | 6.18 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr10_-_93375832 | 6.07 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr7_-_79497940 | 6.05 |
ENSMUST00000107392.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr5_+_87148697 | 5.88 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr5_-_92475927 | 5.84 |
ENSMUST00000113093.5
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr7_-_98010478 | 5.82 |
ENSMUST00000094161.11
ENSMUST00000164726.8 ENSMUST00000206414.2 ENSMUST00000167405.3 |
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr16_-_22848153 | 5.82 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr6_-_6217126 | 5.63 |
ENSMUST00000188414.4
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr11_-_53313950 | 5.53 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr17_-_34000804 | 5.53 |
ENSMUST00000002360.17
|
Angptl4
|
angiopoietin-like 4 |
| chr2_-_27137272 | 5.47 |
ENSMUST00000102886.10
ENSMUST00000129975.2 |
Sardh
|
sarcosine dehydrogenase |
| chr9_+_65536892 | 5.37 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr7_-_98010534 | 5.37 |
ENSMUST00000165257.8
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr6_+_87405968 | 5.17 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr17_+_45997248 | 4.89 |
ENSMUST00000024734.8
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr11_-_75313350 | 4.86 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr8_+_118428643 | 4.85 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr15_+_54274151 | 4.81 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr6_+_120643323 | 4.80 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr6_+_72333209 | 4.78 |
ENSMUST00000206531.2
|
Tmem150a
|
transmembrane protein 150A |
| chr11_-_114952984 | 4.77 |
ENSMUST00000062787.8
|
Cd300e
|
CD300E molecule |
| chr5_+_135754568 | 4.77 |
ENSMUST00000127096.2
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr9_-_110818679 | 4.74 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr11_+_80044931 | 4.53 |
ENSMUST00000021050.14
|
Adap2
|
ArfGAP with dual PH domains 2 |
| chr17_+_84990541 | 4.50 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr17_-_45997046 | 4.49 |
ENSMUST00000143907.3
ENSMUST00000127065.8 |
Tmem63b
|
transmembrane protein 63b |
| chr3_-_131096792 | 4.46 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr11_-_75313412 | 4.44 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr8_+_15107646 | 4.37 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr16_+_14523696 | 4.13 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr12_-_16696958 | 4.10 |
ENSMUST00000238839.2
|
Lpin1
|
lipin 1 |
| chr7_-_45679703 | 4.06 |
ENSMUST00000002850.8
|
Abcc6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr7_-_97066937 | 4.04 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr11_-_109986804 | 3.97 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr5_+_139375540 | 3.96 |
ENSMUST00000100514.3
|
Gpr146
|
G protein-coupled receptor 146 |
| chr15_+_31225302 | 3.95 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr13_-_4329421 | 3.91 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr14_+_31363004 | 3.91 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr5_+_135703426 | 3.90 |
ENSMUST00000153515.8
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr12_-_58315949 | 3.90 |
ENSMUST00000062254.4
|
Clec14a
|
C-type lectin domain family 14, member a |
| chr9_-_110819639 | 3.82 |
ENSMUST00000198702.2
|
Rtp3
|
receptor transporter protein 3 |
| chr16_-_56984137 | 3.78 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chr7_+_24286968 | 3.63 |
ENSMUST00000077191.7
|
Ethe1
|
ethylmalonic encephalopathy 1 |
| chr18_-_12995261 | 3.61 |
ENSMUST00000234427.2
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr1_-_170804116 | 3.61 |
ENSMUST00000159969.8
|
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr19_+_34268053 | 3.60 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr15_+_31568937 | 3.60 |
ENSMUST00000162532.8
ENSMUST00000070918.14 |
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr16_+_26400454 | 3.49 |
ENSMUST00000096129.9
ENSMUST00000166294.9 ENSMUST00000174202.8 ENSMUST00000023156.13 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr7_-_79497922 | 3.49 |
ENSMUST00000205502.2
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr11_-_109986763 | 3.46 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr18_+_84869456 | 3.44 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr17_-_45997132 | 3.38 |
ENSMUST00000113523.9
|
Tmem63b
|
transmembrane protein 63b |
| chr17_-_33166346 | 3.37 |
ENSMUST00000139353.8
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr19_-_29025233 | 3.34 |
ENSMUST00000025696.5
|
Ak3
|
adenylate kinase 3 |
| chr17_-_45903188 | 3.34 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr4_+_60003438 | 3.32 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr4_+_140970161 | 3.30 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr9_-_20887967 | 3.18 |
ENSMUST00000214218.2
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr8_-_41494890 | 3.17 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr1_+_87998487 | 3.16 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr18_-_12995395 | 3.15 |
ENSMUST00000121888.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr17_+_35658131 | 3.09 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr11_+_95733489 | 3.09 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr8_+_56747613 | 3.07 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr2_-_65069383 | 3.06 |
ENSMUST00000155916.8
ENSMUST00000156643.2 |
Cobll1
|
Cobl-like 1 |
| chr18_-_56695333 | 3.04 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_-_180021039 | 3.04 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr16_+_95059121 | 3.03 |
ENSMUST00000113858.3
|
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr19_+_34268071 | 3.01 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr11_+_121325739 | 3.00 |
ENSMUST00000026175.9
ENSMUST00000092302.11 ENSMUST00000103014.4 |
Fn3k
|
fructosamine 3 kinase |
| chr5_+_137348363 | 3.00 |
ENSMUST00000061244.15
|
Ephb4
|
Eph receptor B4 |
| chr9_-_36708599 | 2.97 |
ENSMUST00000238932.2
ENSMUST00000115086.13 |
Ei24
|
etoposide induced 2.4 mRNA |
| chr14_-_123150497 | 2.93 |
ENSMUST00000162164.2
ENSMUST00000110679.9 ENSMUST00000161322.3 ENSMUST00000038075.12 |
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr6_+_116241146 | 2.83 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chrX_+_102365004 | 2.82 |
ENSMUST00000033689.3
|
Cdx4
|
caudal type homeobox 4 |
| chr4_+_152093260 | 2.82 |
ENSMUST00000097773.4
|
Klhl21
|
kelch-like 21 |
| chr9_+_108174052 | 2.82 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr7_-_141850806 | 2.73 |
ENSMUST00000084413.7
ENSMUST00000188274.2 |
Krtap5-1
|
keratin associated protein 5-1 |
| chr1_+_172777976 | 2.73 |
ENSMUST00000215254.2
|
Olfr16
|
olfactory receptor 16 |
| chr7_+_45055077 | 2.72 |
ENSMUST00000107774.3
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr11_+_49500090 | 2.70 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr16_+_95058895 | 2.69 |
ENSMUST00000113859.8
ENSMUST00000152516.2 |
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_-_45996899 | 2.68 |
ENSMUST00000145873.8
|
Tmem63b
|
transmembrane protein 63b |
| chr11_+_118319319 | 2.66 |
ENSMUST00000017590.9
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr14_-_20231871 | 2.63 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr17_-_33166362 | 2.62 |
ENSMUST00000234083.2
ENSMUST00000075253.13 |
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chrX_+_138511360 | 2.62 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr16_-_36810810 | 2.60 |
ENSMUST00000075869.13
|
Fbxo40
|
F-box protein 40 |
| chr8_+_84267722 | 2.57 |
ENSMUST00000058609.5
|
Olfr370
|
olfactory receptor 370 |
| chr3_+_121220146 | 2.56 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr15_-_75760602 | 2.55 |
ENSMUST00000184858.2
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr6_+_54017063 | 2.55 |
ENSMUST00000127323.3
|
Chn2
|
chimerin 2 |
| chr19_+_32734884 | 2.53 |
ENSMUST00000013807.8
|
Pten
|
phosphatase and tensin homolog |
| chr11_+_72098363 | 2.45 |
ENSMUST00000021158.4
|
Txndc17
|
thioredoxin domain containing 17 |
| chr7_-_104979485 | 2.43 |
ENSMUST00000061920.5
|
Olfr690
|
olfactory receptor 690 |
| chr11_-_102298281 | 2.41 |
ENSMUST00000107098.8
ENSMUST00000018821.9 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr4_-_44066960 | 2.37 |
ENSMUST00000173234.8
ENSMUST00000173274.2 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr6_+_17463748 | 2.37 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr2_+_68966125 | 2.33 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr11_-_102298141 | 2.32 |
ENSMUST00000149777.8
ENSMUST00000154001.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr11_-_102297590 | 2.31 |
ENSMUST00000155104.8
ENSMUST00000130436.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr2_-_10085133 | 2.22 |
ENSMUST00000145530.8
ENSMUST00000026887.14 ENSMUST00000114896.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr2_+_144435974 | 2.21 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr10_-_89457115 | 2.20 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr5_-_38649291 | 2.20 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr2_-_26096547 | 2.20 |
ENSMUST00000028302.8
|
Lhx3
|
LIM homeobox protein 3 |
| chr11_-_55499014 | 2.16 |
ENSMUST00000102716.10
|
Glra1
|
glycine receptor, alpha 1 subunit |
| chr18_+_45402018 | 2.15 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr8_+_71195681 | 2.14 |
ENSMUST00000224874.2
|
Pde4c
|
phosphodiesterase 4C, cAMP specific |
| chr4_+_148085179 | 2.14 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr9_+_39933648 | 2.11 |
ENSMUST00000059859.5
|
Olfr981
|
olfactory receptor 981 |
| chr9_-_20888054 | 2.07 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr9_+_107464841 | 2.06 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr8_-_86091970 | 2.05 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_+_25933632 | 2.03 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_+_89808124 | 2.00 |
ENSMUST00000061830.2
|
Olfr1260
|
olfactory receptor 1260 |
| chr7_-_63588610 | 1.99 |
ENSMUST00000063694.10
|
Klf13
|
Kruppel-like factor 13 |
| chr11_-_55498559 | 1.99 |
ENSMUST00000108853.8
ENSMUST00000075603.5 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr4_-_44073016 | 1.98 |
ENSMUST00000128439.8
ENSMUST00000140724.3 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr9_-_45817666 | 1.97 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr7_-_29935150 | 1.96 |
ENSMUST00000189482.2
|
Ovol3
|
ovo like zinc finger 3 |
| chr7_+_114344920 | 1.96 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr6_-_42515954 | 1.96 |
ENSMUST00000171307.2
|
Olfr455
|
olfactory receptor 455 |
| chr11_-_59466995 | 1.93 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr7_-_126046814 | 1.92 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr1_-_53391778 | 1.91 |
ENSMUST00000236737.2
ENSMUST00000027264.10 ENSMUST00000123519.9 |
Gm50478
Asnsd1
|
predicted gene, 50478 asparagine synthetase domain containing 1 |
| chr11_-_102255999 | 1.90 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr3_+_107137924 | 1.85 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr12_-_30423356 | 1.84 |
ENSMUST00000021004.14
|
Sntg2
|
syntrophin, gamma 2 |
| chr2_+_103953083 | 1.83 |
ENSMUST00000040374.6
|
A930018P22Rik
|
RIKEN cDNA A930018P22 gene |
| chr4_-_133066549 | 1.80 |
ENSMUST00000105906.2
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr8_+_110835424 | 1.80 |
ENSMUST00000219454.2
|
Tle7
|
TLE family member 7 |
| chr7_-_44711075 | 1.79 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr12_-_103979900 | 1.78 |
ENSMUST00000058464.5
|
Serpina9
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr7_-_99344779 | 1.78 |
ENSMUST00000137914.2
ENSMUST00000207090.2 ENSMUST00000208225.2 |
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr2_+_143757193 | 1.75 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr7_-_44711130 | 1.74 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr5_+_135382149 | 1.74 |
ENSMUST00000111180.9
ENSMUST00000065785.4 |
Trim50
|
tripartite motif-containing 50 |
| chr4_-_115875055 | 1.73 |
ENSMUST00000049095.6
|
Faah
|
fatty acid amide hydrolase |
| chr6_-_90055488 | 1.73 |
ENSMUST00000203791.3
ENSMUST00000226368.2 |
Vmn1r49
|
vomeronasal 1, receptor 49 |
| chr5_-_93633205 | 1.71 |
ENSMUST00000179091.5
|
Pramel33
|
PRAME like 33 |
| chr8_-_19164846 | 1.70 |
ENSMUST00000062113.8
|
Defb12
|
defensin beta 12 |
| chr7_-_126497421 | 1.70 |
ENSMUST00000121532.8
ENSMUST00000032926.12 |
Tmem219
|
transmembrane protein 219 |
| chr11_+_118319029 | 1.68 |
ENSMUST00000124861.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr4_-_149569614 | 1.68 |
ENSMUST00000126896.2
ENSMUST00000105693.2 ENSMUST00000030845.13 |
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr3_+_127426783 | 1.68 |
ENSMUST00000029587.9
|
Neurog2
|
neurogenin 2 |
| chr16_-_58857120 | 1.67 |
ENSMUST00000206428.2
|
Olfr187
|
olfactory receptor 187 |
| chr3_-_88366159 | 1.66 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_+_164158651 | 1.65 |
ENSMUST00000017144.3
|
Svs6
|
seminal vesicle secretory protein 6 |
| chr5_-_137101108 | 1.65 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr4_-_133066594 | 1.65 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr18_-_78249612 | 1.64 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr9_+_109661817 | 1.63 |
ENSMUST00000200005.5
ENSMUST00000035053.12 ENSMUST00000200345.5 |
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr9_-_58462720 | 1.62 |
ENSMUST00000165365.3
|
Cd276
|
CD276 antigen |
| chr10_+_79690452 | 1.62 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 9.2 | 27.6 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 6.4 | 19.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 5.7 | 17.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 3.0 | 23.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 2.4 | 16.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 2.3 | 9.0 | GO:1901630 | negative regulation of presynaptic membrane organization(GO:1901630) |
| 2.2 | 8.7 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 2.1 | 6.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.9 | 11.6 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.9 | 11.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.9 | 11.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.8 | 5.5 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 1.6 | 13.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.6 | 13.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 1.6 | 11.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.5 | 4.6 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 1.4 | 4.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.2 | 6.1 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.2 | 7.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 1.2 | 7.0 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 1.1 | 5.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.0 | 9.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 1.0 | 46.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 1.0 | 14.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.9 | 5.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.9 | 3.6 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) negative regulation of hypersensitivity(GO:0002884) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.9 | 12.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.8 | 9.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.8 | 3.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.8 | 2.3 | GO:2000642 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.8 | 5.3 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.7 | 5.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.7 | 3.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.6 | 8.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.6 | 3.8 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.6 | 5.7 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.6 | 6.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.6 | 5.9 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.6 | 9.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 3.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.5 | 1.6 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.5 | 2.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.5 | 3.8 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 4.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.5 | 2.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.5 | 4.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.5 | 3.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.5 | 8.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.5 | 17.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.5 | 9.5 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.4 | 1.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 1.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.4 | 7.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.4 | 1.2 | GO:0046144 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 3.5 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.4 | 0.7 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 | 1.1 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.4 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 4.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 1.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 | 0.9 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.3 | 12.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 1.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 4.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.3 | 1.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 3.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.3 | 1.1 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.3 | 3.6 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.3 | 1.9 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.3 | 1.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 0.8 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 3.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 3.9 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.3 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 8.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.3 | 4.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 4.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 5.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 1.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.2 | 0.7 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.2 | 1.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 2.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 2.7 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.2 | 1.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 0.8 | GO:0015840 | urea transport(GO:0015840) |
| 0.2 | 1.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 6.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.2 | 2.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 2.0 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 1.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 0.7 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.2 | 3.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 1.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 1.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 0.6 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.2 | 2.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 3.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.5 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.2 | 0.5 | GO:0033121 | regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.1 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 7.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.3 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 1.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.5 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 3.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 6.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.5 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.1 | 6.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 2.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 1.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.7 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 2.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 3.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.7 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 1.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 3.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 2.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.4 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 1.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 2.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 3.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 2.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.8 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 2.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.2 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 6.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 0.8 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 2.8 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.1 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 2.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.6 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 2.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 1.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 4.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.6 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 1.3 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 4.7 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 0.4 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 1.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 1.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 2.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.8 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 11.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 44.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.8 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 2.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 3.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 2.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 25.6 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.8 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0090537 | CERF complex(GO:0090537) |
| 1.4 | 19.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.1 | 15.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 1.1 | 9.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 11.6 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.7 | 6.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.5 | 1.5 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.5 | 1.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.5 | 7.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.4 | 3.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 1.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.4 | 1.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.4 | 2.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 2.8 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 2.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 1.4 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.3 | 1.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 71.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 0.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 0.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 2.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 1.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 3.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 0.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 1.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 24.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 4.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 0.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 0.5 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.2 | 1.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 2.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 1.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 16.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 20.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 4.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 3.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 4.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 2.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 2.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 26.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 16.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 11.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 7.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 24.3 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 53.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 3.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 9.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 32.7 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 6.9 | 27.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 4.0 | 27.8 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 3.9 | 11.8 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 3.9 | 11.6 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 3.4 | 17.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 3.0 | 9.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 2.9 | 23.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 2.3 | 7.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 2.2 | 8.7 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 2.2 | 6.5 | GO:0046911 | metal chelating activity(GO:0046911) |
| 2.1 | 6.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 1.9 | 13.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.8 | 5.5 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 1.7 | 6.6 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 1.6 | 42.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.4 | 16.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.1 | 5.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.1 | 3.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.0 | 5.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 8.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.9 | 3.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.9 | 3.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.9 | 4.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.9 | 8.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.8 | 2.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.8 | 4.2 | GO:0030977 | taurine binding(GO:0030977) |
| 0.8 | 3.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.8 | 4.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.7 | 8.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 6.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 5.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 3.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.6 | 4.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.6 | 14.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 11.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.5 | 9.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.5 | 2.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.5 | 1.9 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.5 | 11.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.4 | 3.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 1.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.4 | 5.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.4 | 11.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 3.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 11.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.4 | 3.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 2.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.4 | 3.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.4 | 1.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.4 | 2.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 0.7 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.3 | 3.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 5.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 51.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 1.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 0.9 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.3 | 0.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 1.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 4.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 2.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 5.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 0.8 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.2 | 2.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 9.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 4.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 3.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 1.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 4.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 6.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 0.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 4.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 3.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 4.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 3.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 2.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 3.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 6.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.6 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.8 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.9 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.1 | 0.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 1.2 | GO:0015250 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.1 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 13.4 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.1 | 2.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 2.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 5.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 3.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.2 | GO:0005501 | retinoid binding(GO:0005501) |
| 0.0 | 0.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 5.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 41.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 3.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 3.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 2.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 4.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 9.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 6.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 23.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 2.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 5.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 75.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 8.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 7.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 5.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 6.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 16.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 6.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 3.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 50.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.3 | 22.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.2 | 28.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.9 | 30.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.8 | 7.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.7 | 11.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.6 | 16.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.5 | 11.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 8.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 6.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 9.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 2.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 3.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 6.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 13.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 4.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 2.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 5.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 2.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 11.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 8.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 3.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 5.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 3.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 5.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 2.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |