Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zbtb3
Z-value: 0.98
Transcription factors associated with Zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb3
|
ENSMUSG00000071661.8 | Zbtb3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb3 | mm39_v1_chr19_+_8779903_8779922 | -0.51 | 5.7e-06 | Click! |
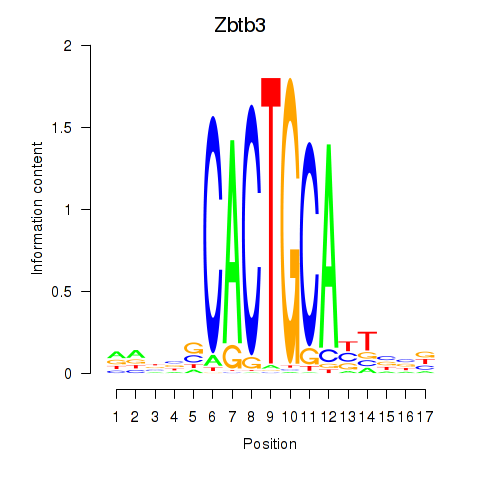
Activity profile of Zbtb3 motif
Sorted Z-values of Zbtb3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_53994813 | 8.56 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr14_+_53872276 | 7.32 |
ENSMUST00000103658.4
|
Trav13-2
|
T cell receptor alpha variable 13-2 |
| chr14_+_53995108 | 6.84 |
ENSMUST00000184905.2
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr8_-_84769170 | 6.27 |
ENSMUST00000005601.9
|
Il27ra
|
interleukin 27 receptor, alpha |
| chr14_+_54032814 | 6.12 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr14_+_53180221 | 5.94 |
ENSMUST00000197954.2
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr11_-_120622770 | 5.74 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr15_-_78657640 | 5.53 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_162687369 | 5.37 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr1_-_162687254 | 4.94 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr4_-_150093435 | 4.77 |
ENSMUST00000030830.4
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr14_+_53088747 | 4.60 |
ENSMUST00000103588.4
|
Trav13d-1
|
T cell receptor alpha variable 13D-1 |
| chr14_+_53574579 | 4.36 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr14_+_53399856 | 4.07 |
ENSMUST00000198359.2
|
Trav13n-1
|
T cell receptor alpha variable 13N-1 |
| chr14_+_53601293 | 3.94 |
ENSMUST00000103634.3
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr14_+_53180476 | 3.94 |
ENSMUST00000103596.3
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr14_+_53539493 | 3.87 |
ENSMUST00000103627.3
|
Trav4n-4
|
T cell receptor alpha variable 4N-4 |
| chr10_-_53951825 | 3.86 |
ENSMUST00000003843.16
|
Man1a
|
mannosidase 1, alpha |
| chr1_-_162687488 | 3.77 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_-_25517945 | 3.69 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr18_+_44467133 | 3.67 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr1_+_156138286 | 3.61 |
ENSMUST00000027896.10
|
Nphs2
|
nephrosis 2, podocin |
| chr6_-_136834725 | 3.48 |
ENSMUST00000032341.3
|
Art4
|
ADP-ribosyltransferase 4 |
| chr10_+_60182630 | 3.39 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr15_-_66985760 | 3.37 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr14_+_53491249 | 3.33 |
ENSMUST00000196941.2
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr8_-_96615138 | 3.30 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr10_-_53951796 | 3.21 |
ENSMUST00000105470.9
|
Man1a
|
mannosidase 1, alpha |
| chr9_+_46180362 | 3.11 |
ENSMUST00000214202.2
ENSMUST00000215458.2 ENSMUST00000215187.2 ENSMUST00000213878.2 ENSMUST00000034584.4 |
Apoa5
|
apolipoprotein A-V |
| chr12_+_112455882 | 3.05 |
ENSMUST00000057465.7
ENSMUST00000223266.2 |
A530016L24Rik
|
RIKEN cDNA A530016L24 gene |
| chr19_-_8382424 | 3.04 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr14_+_30856687 | 3.03 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr12_-_113552322 | 2.80 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr14_+_53782432 | 2.74 |
ENSMUST00000103651.4
|
Trav13-1
|
T cell receptor alpha variable 13-1 |
| chr14_+_53310220 | 2.67 |
ENSMUST00000196079.2
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chrX_+_106192510 | 2.64 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr14_+_53836282 | 2.58 |
ENSMUST00000103655.3
|
Trav4-3
|
T cell receptor alpha variable 4-3 |
| chrX_-_161747552 | 2.57 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr14_+_53137426 | 2.51 |
ENSMUST00000103592.2
|
Trav4d-3
|
T cell receptor alpha variable 4D-3 |
| chr9_+_92157655 | 2.45 |
ENSMUST00000034932.14
ENSMUST00000180154.8 |
Plscr2
|
phospholipid scramblase 2 |
| chr14_+_53270305 | 2.35 |
ENSMUST00000179512.3
|
Trav13d-3
|
T cell receptor alpha variable 13D-3 |
| chr6_+_72281587 | 2.32 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr14_+_53491504 | 2.32 |
ENSMUST00000103622.3
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr6_-_128868068 | 2.11 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr10_+_128104525 | 2.11 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr9_+_44990447 | 2.06 |
ENSMUST00000050020.8
|
Jaml
|
junction adhesion molecule like |
| chr11_+_51858476 | 2.04 |
ENSMUST00000102763.5
|
Cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chrX_-_153999440 | 1.96 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr14_+_53310461 | 1.88 |
ENSMUST00000103607.3
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chr1_+_132973724 | 1.88 |
ENSMUST00000077730.7
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chrX_-_153999333 | 1.83 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr4_-_40853950 | 1.83 |
ENSMUST00000030121.13
ENSMUST00000108096.3 |
B4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr11_+_6511133 | 1.81 |
ENSMUST00000160633.8
ENSMUST00000109721.3 |
Ccm2
|
cerebral cavernous malformation 2 |
| chr9_+_21746785 | 1.80 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr9_+_44410417 | 1.75 |
ENSMUST00000074989.7
ENSMUST00000218913.2 |
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr14_+_53878158 | 1.73 |
ENSMUST00000179267.4
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr9_+_44990502 | 1.72 |
ENSMUST00000216426.2
|
Jaml
|
junction adhesion molecule like |
| chr9_+_92157799 | 1.70 |
ENSMUST00000126911.2
|
Plscr2
|
phospholipid scramblase 2 |
| chrX_-_166047275 | 1.61 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr11_-_48793009 | 1.57 |
ENSMUST00000109212.3
|
Gm5431
|
predicted gene 5431 |
| chr1_-_133849131 | 1.55 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr15_-_89012838 | 1.52 |
ENSMUST00000082197.12
|
Hdac10
|
histone deacetylase 10 |
| chr7_+_45084300 | 1.52 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr15_-_79326311 | 1.51 |
ENSMUST00000230942.2
|
Csnk1e
|
casein kinase 1, epsilon |
| chr6_+_124690060 | 1.49 |
ENSMUST00000130279.2
|
Phb2
|
prohibitin 2 |
| chr7_-_141023902 | 1.49 |
ENSMUST00000026580.12
|
Pidd1
|
p53 induced death domain protein 1 |
| chr14_+_36789999 | 1.46 |
ENSMUST00000057176.5
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr14_+_53878403 | 1.39 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr9_+_66853343 | 1.39 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr19_-_8109346 | 1.37 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr17_+_35235552 | 1.36 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chrX_-_156351979 | 1.35 |
ENSMUST00000065806.5
|
Yy2
|
Yy2 transcription factor |
| chr3_-_37286714 | 1.34 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr10_+_61531282 | 1.33 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr9_+_107458495 | 1.31 |
ENSMUST00000040059.9
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr12_-_101785307 | 1.29 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr7_-_119744509 | 1.27 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr8_+_84441854 | 1.25 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a |
| chr11_-_48792973 | 1.23 |
ENSMUST00000109210.8
|
Gm5431
|
predicted gene 5431 |
| chr2_-_127634387 | 1.22 |
ENSMUST00000135091.2
|
Mtln
|
mitoregulin |
| chr10_-_62723238 | 1.22 |
ENSMUST00000228901.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr7_-_80037622 | 1.18 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr3_+_82962823 | 1.16 |
ENSMUST00000150268.8
ENSMUST00000122128.2 |
Plrg1
|
pleiotropic regulator 1 |
| chr17_+_24768808 | 1.16 |
ENSMUST00000228550.2
ENSMUST00000035565.5 |
Pkd1
|
polycystin 1, transient receptor poteintial channel interacting |
| chr4_+_40269563 | 1.16 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr15_+_78128990 | 1.15 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr9_-_121620150 | 1.13 |
ENSMUST00000215910.2
ENSMUST00000215477.2 ENSMUST00000163981.3 |
Hhatl
|
hedgehog acyltransferase-like |
| chr3_+_107198528 | 1.13 |
ENSMUST00000029502.14
|
Slc16a4
|
solute carrier family 16 (monocarboxylic acid transporters), member 4 |
| chr2_+_152404897 | 1.11 |
ENSMUST00000238626.2
|
Gm17416
|
predicted gene, 17416 |
| chr2_-_35226981 | 1.09 |
ENSMUST00000028241.7
|
Stom
|
stomatin |
| chr7_-_80037688 | 1.09 |
ENSMUST00000080932.8
|
Fes
|
feline sarcoma oncogene |
| chr6_+_134897364 | 1.09 |
ENSMUST00000067327.11
ENSMUST00000003115.9 |
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr4_-_49473904 | 1.06 |
ENSMUST00000135976.2
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr11_+_105956867 | 1.05 |
ENSMUST00000002048.8
|
Taco1
|
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr2_-_51862941 | 1.03 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr7_+_28050077 | 1.00 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr11_-_16952929 | 0.99 |
ENSMUST00000156101.2
|
Plek
|
pleckstrin |
| chr10_-_62723103 | 0.98 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr3_+_107198618 | 0.98 |
ENSMUST00000106723.2
|
Slc16a4
|
solute carrier family 16 (monocarboxylic acid transporters), member 4 |
| chr12_-_101784727 | 0.98 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr7_-_44497950 | 0.96 |
ENSMUST00000208890.2
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chr7_+_101555111 | 0.94 |
ENSMUST00000033131.12
ENSMUST00000193465.2 |
Lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr18_-_52662728 | 0.90 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chrX_-_72380446 | 0.89 |
ENSMUST00000207943.2
|
Gm45015
|
predicted gene 45015 |
| chr19_-_58444336 | 0.85 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr7_+_45084257 | 0.84 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr7_-_121700958 | 0.84 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr7_+_97049210 | 0.81 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chrX_-_166047289 | 0.81 |
ENSMUST00000133722.2
|
Tlr8
|
toll-like receptor 8 |
| chr7_-_30826376 | 0.80 |
ENSMUST00000098548.8
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr15_+_78129040 | 0.70 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr7_-_110681402 | 0.69 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr1_-_171108754 | 0.68 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr11_+_116424082 | 0.67 |
ENSMUST00000154034.8
|
Sphk1
|
sphingosine kinase 1 |
| chr11_+_77654072 | 0.66 |
ENSMUST00000108375.9
|
Myo18a
|
myosin XVIIIA |
| chr16_+_58969192 | 0.63 |
ENSMUST00000075381.3
|
Olfr195
|
olfactory receptor 195 |
| chr17_+_48666919 | 0.62 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr16_+_11223512 | 0.62 |
ENSMUST00000096273.9
|
Snx29
|
sorting nexin 29 |
| chr10_+_69932930 | 0.61 |
ENSMUST00000147545.8
|
Ccdc6
|
coiled-coil domain containing 6 |
| chr8_+_84441806 | 0.61 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr8_+_84442133 | 0.60 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr11_+_73067909 | 0.60 |
ENSMUST00000040687.12
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr13_+_19362068 | 0.59 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr6_-_58418303 | 0.58 |
ENSMUST00000228577.2
ENSMUST00000227466.2 |
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr13_+_96679110 | 0.57 |
ENSMUST00000179226.8
|
Cert1
|
ceramide transporter 1 |
| chr3_-_115800989 | 0.57 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr9_-_57342242 | 0.56 |
ENSMUST00000215961.2
|
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chrX_-_36990581 | 0.55 |
ENSMUST00000170643.4
ENSMUST00000089062.8 |
Rhox9
|
reproductive homeobox 9 |
| chr2_+_172235820 | 0.55 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr14_+_66378382 | 0.53 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr11_-_102076028 | 0.52 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr2_+_74566740 | 0.52 |
ENSMUST00000111982.8
|
Hoxd3
|
homeobox D3 |
| chr11_+_73068063 | 0.51 |
ENSMUST00000108477.2
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr4_-_43700807 | 0.48 |
ENSMUST00000055545.5
|
Olfr70
|
olfactory receptor 70 |
| chr19_-_34855242 | 0.47 |
ENSMUST00000238065.2
|
Pank1
|
pantothenate kinase 1 |
| chr8_-_69187708 | 0.47 |
ENSMUST00000136060.8
ENSMUST00000130214.8 ENSMUST00000078350.13 |
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr16_-_58710581 | 0.47 |
ENSMUST00000058564.4
|
Olfr178
|
olfactory receptor 178 |
| chr19_-_34855278 | 0.46 |
ENSMUST00000112460.3
|
Pank1
|
pantothenate kinase 1 |
| chr7_+_138448061 | 0.44 |
ENSMUST00000041097.13
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr11_-_5691117 | 0.43 |
ENSMUST00000140922.2
ENSMUST00000093362.12 |
Urgcp
|
upregulator of cell proliferation |
| chr18_+_90528246 | 0.43 |
ENSMUST00000025515.7
ENSMUST00000235708.2 |
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr17_-_37523969 | 0.42 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr11_-_86561980 | 0.41 |
ENSMUST00000143991.3
|
Vmp1
|
vacuole membrane protein 1 |
| chr7_-_44498305 | 0.41 |
ENSMUST00000207293.2
ENSMUST00000207532.2 |
Tbc1d17
|
TBC1 domain family, member 17 |
| chr18_+_90528308 | 0.37 |
ENSMUST00000235634.2
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr19_-_21630143 | 0.37 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr8_+_84682136 | 0.35 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr11_+_115921129 | 0.35 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr9_-_49330736 | 0.34 |
ENSMUST00000216227.2
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr12_+_81906738 | 0.33 |
ENSMUST00000221721.2
ENSMUST00000021567.6 |
Pcnx
|
pecanex homolog |
| chr13_+_96679233 | 0.33 |
ENSMUST00000077672.12
ENSMUST00000109444.3 |
Cert1
|
ceramide transporter 1 |
| chr7_+_28466658 | 0.33 |
ENSMUST00000155327.8
|
Sirt2
|
sirtuin 2 |
| chr9_+_122942280 | 0.33 |
ENSMUST00000026891.5
ENSMUST00000215377.2 |
Exosc7
|
exosome component 7 |
| chr16_+_17798292 | 0.32 |
ENSMUST00000075371.5
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr11_+_109304735 | 0.32 |
ENSMUST00000055404.8
|
9930022D16Rik
|
RIKEN cDNA 9930022D16 gene |
| chr15_+_103181311 | 0.31 |
ENSMUST00000100162.5
ENSMUST00000230893.2 |
Copz1
|
coatomer protein complex, subunit zeta 1 |
| chr4_+_138606671 | 0.31 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr6_-_40590244 | 0.29 |
ENSMUST00000076565.3
|
Tas2r138
|
taste receptor, type 2, member 138 |
| chr2_-_89855921 | 0.28 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr1_-_121255400 | 0.27 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr2_-_180798785 | 0.26 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr16_+_34511073 | 0.25 |
ENSMUST00000231609.2
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr17_-_21125353 | 0.25 |
ENSMUST00000235948.2
ENSMUST00000238096.2 |
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chr2_+_119378178 | 0.24 |
ENSMUST00000014221.13
ENSMUST00000119172.2 |
Chp1
|
calcineurin-like EF hand protein 1 |
| chr2_+_172235702 | 0.24 |
ENSMUST00000099061.9
ENSMUST00000103073.9 |
Cass4
|
Cas scaffolding protein family member 4 |
| chrX_+_36915787 | 0.23 |
ENSMUST00000006362.4
|
Rhox6
|
reproductive homeobox 6 |
| chr16_+_34510918 | 0.23 |
ENSMUST00000023532.7
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr9_+_21527526 | 0.22 |
ENSMUST00000174008.8
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr18_-_80512850 | 0.22 |
ENSMUST00000036229.13
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr7_+_138448308 | 0.21 |
ENSMUST00000155672.8
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr8_-_3517617 | 0.20 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr6_-_28421678 | 0.18 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr5_+_146885450 | 0.17 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr2_-_157179344 | 0.17 |
ENSMUST00000109536.8
|
Ghrh
|
growth hormone releasing hormone |
| chr2_-_51863203 | 0.17 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr1_-_121255753 | 0.15 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr9_-_89505178 | 0.15 |
ENSMUST00000044491.13
|
Minar1
|
membrane integral NOTCH2 associated receptor 1 |
| chr6_-_86503178 | 0.15 |
ENSMUST00000053015.7
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr5_-_137530214 | 0.15 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr13_-_95359543 | 0.14 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr1_+_181952302 | 0.13 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr9_+_21527462 | 0.13 |
ENSMUST00000034707.15
ENSMUST00000098948.10 |
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr16_+_22738987 | 0.12 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr3_+_79793237 | 0.10 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr13_-_96678987 | 0.10 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr11_-_53918916 | 0.09 |
ENSMUST00000020586.7
|
Slc22a4
|
solute carrier family 22 (organic cation transporter), member 4 |
| chr16_+_22739191 | 0.09 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr2_+_140012560 | 0.08 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr7_-_114162125 | 0.08 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr10_+_79766254 | 0.07 |
ENSMUST00000131118.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr18_+_34380738 | 0.05 |
ENSMUST00000066133.7
|
Apc
|
APC, WNT signaling pathway regulator |
| chr12_-_72711509 | 0.05 |
ENSMUST00000221750.2
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr16_+_22739028 | 0.02 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr15_-_77639418 | 0.02 |
ENSMUST00000229445.2
|
Apol8
|
apolipoprotein L 8 |
| chr11_+_58868919 | 0.01 |
ENSMUST00000108809.8
ENSMUST00000108810.10 ENSMUST00000093061.7 |
Trim11
|
tripartite motif-containing 11 |
| chr10_+_79500387 | 0.00 |
ENSMUST00000020554.8
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr6_+_57183497 | 0.00 |
ENSMUST00000227298.2
|
Vmn1r13
|
vomeronasal 1 receptor 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 1.0 | 5.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.8 | 3.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.8 | 3.8 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.7 | 14.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.6 | 5.6 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.5 | 3.4 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 1.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 | 3.6 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.4 | 6.3 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.4 | 1.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.4 | 1.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.3 | 5.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.0 | GO:0010925 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.3 | 1.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 4.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 3.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 0.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.3 | 3.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 2.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 4.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 7.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 1.3 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.2 | 0.7 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.2 | 1.5 | GO:0060744 | positive regulation of exit from mitosis(GO:0031536) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 1.3 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 1.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 0.9 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.7 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.8 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 0.8 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.3 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 1.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 2.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.1 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.4 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 2.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 3.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 1.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 1.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 3.6 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.6 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 2.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 2.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 2.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 1.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 2.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 4.2 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 2.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 2.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 2.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 3.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 3.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 4.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 6.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.2 | GO:0000974 | Prp19 complex(GO:0000974) DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 4.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 4.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.6 | 4.8 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 1.2 | 14.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.1 | 3.3 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 3.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.8 | 5.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.8 | 2.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.7 | 6.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.6 | 1.8 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 7.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.5 | 3.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 3.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 2.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 0.8 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.3 | 0.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 4.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 3.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 2.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 4.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 3.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.8 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.2 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 5.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 6.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 2.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0030957 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) Tat protein binding(GO:0030957) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 4.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 7.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 14.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 4.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 2.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 3.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 2.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |