Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zbtb33_Chd2
Z-value: 1.98
Transcription factors associated with Zbtb33_Chd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
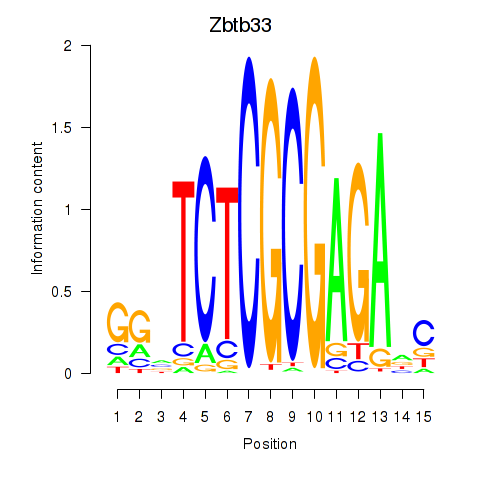
Zbtb33
|
ENSMUSG00000048047.4 | Zbtb33 |
|
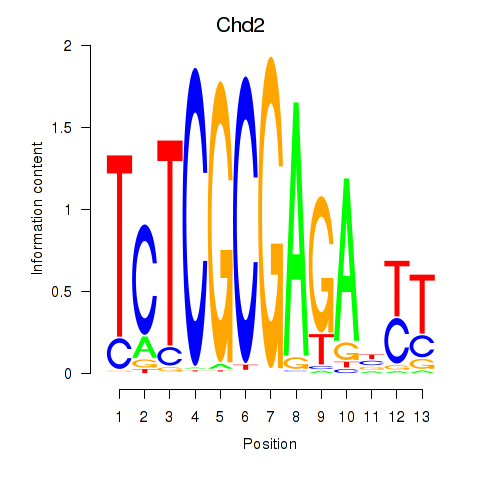
Chd2
|
ENSMUSG00000078671.12 | Chd2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Chd2 | mm39_v1_chr7_-_73187369_73187513 | 0.59 | 4.5e-08 | Click! |
| Zbtb33 | mm39_v1_chrX_+_37278636_37278722 | 0.47 | 3.5e-05 | Click! |
Activity profile of Zbtb33_Chd2 motif
Sorted Z-values of Zbtb33_Chd2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb33_Chd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 1.9 | 14.9 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 1.8 | 17.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.7 | 5.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.5 | 7.6 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 1.5 | 4.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.4 | 15.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.4 | 4.1 | GO:0048822 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 1.3 | 4.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.3 | 9.0 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 1.2 | 7.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 1.1 | 3.4 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 1.1 | 15.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 1.1 | 3.3 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 1.0 | 12.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.9 | 2.8 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.9 | 3.7 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.9 | 3.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.9 | 4.5 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.9 | 9.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.8 | 4.1 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.8 | 2.4 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.8 | 4.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.8 | 3.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.7 | 3.6 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.7 | 2.7 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.7 | 4.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.7 | 2.0 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.7 | 2.0 | GO:0042262 | DNA protection(GO:0042262) |
| 0.7 | 2.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.6 | 5.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.6 | 4.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.6 | 2.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.6 | 4.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.5 | 4.4 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.5 | 5.8 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.5 | 17.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.5 | 2.9 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.5 | 2.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 0.9 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.5 | 15.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.5 | 1.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.5 | 3.7 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.4 | 1.8 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.4 | 3.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 | 1.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 12.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.4 | 1.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.4 | 2.4 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.4 | 1.5 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.4 | 1.9 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.4 | 1.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.4 | 3.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 3.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 3.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 7.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.3 | 5.7 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.3 | 3.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.3 | 9.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 10.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 4.4 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.3 | 1.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 0.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.3 | 6.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 3.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 7.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 2.0 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 5.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 3.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 0.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 2.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 1.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 2.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 2.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 2.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.2 | 1.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 0.6 | GO:1903722 | multivesicular body assembly(GO:0036258) regulation of centriole elongation(GO:1903722) |
| 0.2 | 3.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) |
| 0.2 | 0.6 | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.2 | 1.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 3.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 1.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 1.5 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 3.3 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.2 | 2.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 1.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.2 | 0.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 3.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 0.5 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 7.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 0.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 3.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 5.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 2.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.2 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 0.4 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.4 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 0.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 1.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 14.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 5.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 2.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 3.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 3.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 2.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 3.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.3 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.1 | 1.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 2.9 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 1.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.8 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 1.0 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 1.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.8 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 3.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 8.9 | GO:0031109 | microtubule polymerization or depolymerization(GO:0031109) |
| 0.1 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.5 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 0.5 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.4 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.4 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 2.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.4 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 1.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.8 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 4.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 4.6 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 3.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 1.3 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 5.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.7 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 5.6 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 7.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 1.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 4.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.4 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 0.3 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 8.2 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 1.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.4 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 34.5 | GO:0000796 | condensin complex(GO:0000796) |
| 2.7 | 8.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.4 | 9.5 | GO:0035101 | FACT complex(GO:0035101) |
| 1.8 | 12.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 1.8 | 8.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.3 | 19.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.1 | 5.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.1 | 5.4 | GO:0031251 | PAN complex(GO:0031251) |
| 1.1 | 6.4 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.0 | 3.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 1.0 | 3.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.7 | 2.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.7 | 2.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.7 | 6.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.6 | 3.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.6 | 2.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 4.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.5 | 4.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.5 | 3.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 3.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.5 | 2.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 7.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.4 | 5.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 1.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.4 | 15.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.4 | 3.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 2.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 2.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 19.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 3.7 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.3 | 4.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 2.4 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 5.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 3.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 9.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 2.9 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 9.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 3.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 17.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 0.9 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 7.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.2 | 1.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 3.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 5.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 1.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.2 | 2.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 1.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 0.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 1.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.6 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 2.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 3.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 1.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 2.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.7 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.9 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 3.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 5.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 2.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 3.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 6.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 19.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.4 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 3.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.7 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.7 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 4.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 3.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 3.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 24.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 6.0 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 4.2 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 8.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 7.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.3 | 4.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.3 | 7.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.3 | 5.0 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 0.8 | 6.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.8 | 8.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.8 | 5.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.7 | 3.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.7 | 2.0 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.6 | 5.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.6 | 12.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.6 | 2.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.6 | 2.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.5 | 2.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.5 | 4.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 2.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 3.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 1.8 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.4 | 1.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.4 | 1.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.4 | 4.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 2.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.4 | 3.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.4 | 1.1 | GO:0045142 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.4 | 2.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.4 | 1.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 2.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 2.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 5.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 3.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 3.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 1.2 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 0.3 | 2.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.3 | 1.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 5.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.3 | 2.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.3 | 3.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 3.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 3.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 1.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 17.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 0.7 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 5.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 2.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 5.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 10.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 0.8 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 22.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 1.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 6.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 2.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.2 | 3.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 2.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 26.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 2.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 3.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 1.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 3.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 3.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 4.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 7.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 4.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 3.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 4.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 3.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 17.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 5.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 7.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.4 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 3.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 2.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 1.0 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 4.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 6.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 7.9 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 10.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 6.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.8 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 1.1 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 31.7 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 7.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 38.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 21.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 6.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 9.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 5.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 4.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 4.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 7.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 4.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 9.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 5.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 4.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.6 | 10.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.5 | 9.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 5.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 47.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.3 | 34.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 6.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 6.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 2.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 7.0 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 7.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 4.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 4.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 5.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 7.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 6.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 4.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 5.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 3.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.1 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.1 | 4.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 2.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 6.4 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 2.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |