Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
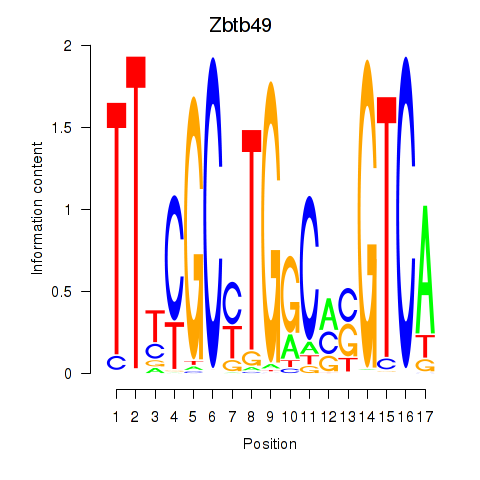
Results for Zbtb49
Z-value: 0.49
Transcription factors associated with Zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb49
|
ENSMUSG00000029127.16 | Zbtb49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb49 | mm39_v1_chr5_-_38377746_38377801 | -0.37 | 1.4e-03 | Click! |
Activity profile of Zbtb49 motif
Sorted Z-values of Zbtb49 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb49
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_71176811 | 10.55 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr19_-_39637489 | 7.44 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr15_-_34495329 | 6.13 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr7_-_139734637 | 3.57 |
ENSMUST00000059241.8
|
Sprn
|
shadow of prion protein |
| chr17_+_24955613 | 3.10 |
ENSMUST00000115262.9
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr17_+_24955647 | 3.01 |
ENSMUST00000101800.7
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr18_-_62044871 | 2.96 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr7_-_34914675 | 2.19 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr3_+_154302989 | 1.67 |
ENSMUST00000140644.8
ENSMUST00000144764.8 ENSMUST00000155232.2 |
Cryz
|
crystallin, zeta |
| chr12_+_89779237 | 1.62 |
ENSMUST00000110133.9
ENSMUST00000110130.4 |
Nrxn3
|
neurexin III |
| chr9_+_40180497 | 1.47 |
ENSMUST00000049941.12
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr9_+_40180569 | 1.45 |
ENSMUST00000176185.8
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr9_+_40180726 | 1.40 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr12_+_89779178 | 1.31 |
ENSMUST00000238943.2
|
Nrxn3
|
neurexin III |
| chr2_-_80411724 | 1.25 |
ENSMUST00000111760.3
|
Nckap1
|
NCK-associated protein 1 |
| chr2_-_80411578 | 1.02 |
ENSMUST00000028386.12
|
Nckap1
|
NCK-associated protein 1 |
| chr5_+_35971697 | 0.99 |
ENSMUST00000130233.8
|
Ablim2
|
actin-binding LIM protein 2 |
| chr5_-_108777578 | 0.90 |
ENSMUST00000046603.15
|
Gak
|
cyclin G associated kinase |
| chr15_-_100320872 | 0.80 |
ENSMUST00000138843.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr2_+_131048998 | 0.75 |
ENSMUST00000153097.3
|
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr7_-_97848688 | 0.65 |
ENSMUST00000098278.4
|
B3gnt6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr15_-_3333003 | 0.63 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr1_+_32211792 | 0.57 |
ENSMUST00000027226.12
ENSMUST00000189878.2 ENSMUST00000188257.7 ENSMUST00000185666.2 |
Khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chrX_+_20714782 | 0.54 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chr8_+_105727456 | 0.53 |
ENSMUST00000172032.4
|
Ces2h
|
carboxylesterase 2H |
| chr15_+_100321074 | 0.51 |
ENSMUST00000148928.2
|
Gm5475
|
predicted gene 5475 |
| chr18_+_23886765 | 0.40 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_+_45341589 | 0.38 |
ENSMUST00000239471.2
ENSMUST00000034592.11 ENSMUST00000239429.2 |
Dscaml1
|
DS cell adhesion molecule like 1 |
| chr14_+_53279810 | 0.26 |
ENSMUST00000198439.5
ENSMUST00000103605.3 |
Trav8d-2
|
T cell receptor alpha variable 8D-2 |
| chr15_+_34495441 | 0.02 |
ENSMUST00000052290.14
ENSMUST00000079028.6 |
Pop1
|
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.7 | 10.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.4 | 4.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.3 | 2.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 0.9 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 7.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 2.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.5 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 1.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 6.1 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 3.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 4.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 6.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 7.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.0 | 6.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.7 | 6.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.5 | 4.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 1.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 7.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 2.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 4.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 10.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |