Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
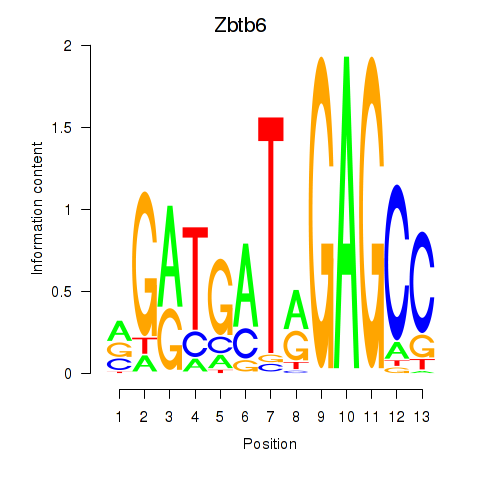
Results for Zbtb6
Z-value: 1.31
Transcription factors associated with Zbtb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb6
|
ENSMUSG00000066798.4 | Zbtb6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb6 | mm39_v1_chr2_-_37333109_37333210 | 0.20 | 9.5e-02 | Click! |
Activity profile of Zbtb6 motif
Sorted Z-values of Zbtb6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_60806810 | 8.96 |
ENSMUST00000163779.8
|
Snca
|
synuclein, alpha |
| chr9_-_99599312 | 8.50 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr2_-_126460575 | 6.61 |
ENSMUST00000028838.5
|
Hdc
|
histidine decarboxylase |
| chr15_+_78798116 | 5.83 |
ENSMUST00000089378.5
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr11_-_29975916 | 5.17 |
ENSMUST00000058902.6
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr11_-_102787950 | 5.15 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr1_+_75526225 | 4.95 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr15_+_82140224 | 4.92 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr1_+_78488499 | 4.90 |
ENSMUST00000012331.7
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chrX_+_135723531 | 4.55 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chrX_+_135723420 | 4.34 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr11_-_65053710 | 4.32 |
ENSMUST00000093002.12
ENSMUST00000047463.15 |
Arhgap44
|
Rho GTPase activating protein 44 |
| chr6_-_113696390 | 4.31 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr1_+_78488440 | 4.19 |
ENSMUST00000152111.2
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr1_+_134110142 | 4.06 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr5_+_73563418 | 3.97 |
ENSMUST00000031040.13
ENSMUST00000065543.8 |
Cwh43
|
cell wall biogenesis 43 C-terminal homolog |
| chr6_-_113696809 | 3.93 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr5_-_84565218 | 3.88 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr11_-_102787972 | 3.83 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr5_+_30745447 | 3.79 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr14_-_55150547 | 3.79 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr6_+_86005663 | 3.68 |
ENSMUST00000204059.3
|
Add2
|
adducin 2 (beta) |
| chr7_-_144761806 | 3.67 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr2_-_113659360 | 3.55 |
ENSMUST00000024005.8
|
Scg5
|
secretogranin V |
| chr15_+_103411461 | 3.39 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chrX_-_20157966 | 3.38 |
ENSMUST00000115393.3
ENSMUST00000072451.11 |
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr6_+_29917004 | 3.37 |
ENSMUST00000151738.4
ENSMUST00000115224.8 ENSMUST00000046028.12 |
Strip2
|
striatin interacting protein 2 |
| chr17_+_37180437 | 3.36 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr5_-_18565353 | 3.21 |
ENSMUST00000074694.7
|
Gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
| chr9_+_102988940 | 3.21 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr15_-_101621332 | 3.16 |
ENSMUST00000023709.7
|
Krt5
|
keratin 5 |
| chr16_+_17716480 | 3.14 |
ENSMUST00000055374.8
|
Tssk2
|
testis-specific serine kinase 2 |
| chr8_-_86091946 | 3.12 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr15_+_39061612 | 2.94 |
ENSMUST00000082054.12
ENSMUST00000227243.2 ENSMUST00000042917.10 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr17_-_40519480 | 2.94 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr1_+_134109888 | 2.94 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr9_-_62417780 | 2.84 |
ENSMUST00000164246.9
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr6_+_86005729 | 2.79 |
ENSMUST00000203366.3
|
Add2
|
adducin 2 (beta) |
| chr6_+_123239076 | 2.72 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr5_+_137551774 | 2.71 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr2_+_90948481 | 2.71 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr11_-_115158062 | 2.65 |
ENSMUST00000106554.2
|
Grin2c
|
glutamate receptor, ionotropic, NMDA2C (epsilon 3) |
| chr18_+_37637317 | 2.65 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr14_-_55204054 | 2.60 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr8_+_106331866 | 2.55 |
ENSMUST00000043531.10
|
Ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr1_-_75240551 | 2.49 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr5_-_71253107 | 2.46 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr14_-_55204092 | 2.44 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chrX_-_165992311 | 2.41 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr16_+_17095416 | 2.40 |
ENSMUST00000232364.2
|
Tmem191c
|
transmembrane protein 191C |
| chr9_-_61883845 | 2.38 |
ENSMUST00000113990.2
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr12_-_116226565 | 2.37 |
ENSMUST00000039349.8
|
Dync2i1
|
dynein 2 intermediate chain 1 |
| chr6_+_124973644 | 2.35 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr17_-_46798566 | 2.34 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chr13_+_110531571 | 2.31 |
ENSMUST00000022212.9
|
Plk2
|
polo like kinase 2 |
| chr11_-_42073737 | 2.31 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr6_+_124973752 | 2.31 |
ENSMUST00000162000.4
|
Pianp
|
PILR alpha associated neural protein |
| chr8_-_86091970 | 2.30 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_23295603 | 2.26 |
ENSMUST00000163739.3
ENSMUST00000210656.2 |
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr8_+_75820240 | 2.23 |
ENSMUST00000005548.8
|
Hmox1
|
heme oxygenase 1 |
| chr17_-_25394445 | 2.21 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr2_+_177834868 | 2.19 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr7_+_118454957 | 2.18 |
ENSMUST00000208658.2
ENSMUST00000098087.9 ENSMUST00000106547.2 |
Iqck
|
IQ motif containing K |
| chr3_+_92864693 | 2.17 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr14_+_56640038 | 2.17 |
ENSMUST00000095793.3
ENSMUST00000223627.2 |
Rnf17
|
ring finger protein 17 |
| chr9_+_54606832 | 2.15 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr9_+_22322802 | 2.13 |
ENSMUST00000058868.9
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr9_+_54606144 | 2.13 |
ENSMUST00000120452.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr17_-_35069136 | 2.12 |
ENSMUST00000046022.16
|
Skiv2l
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr16_+_17712061 | 2.07 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr2_+_21372338 | 2.06 |
ENSMUST00000055946.8
|
Gpr158
|
G protein-coupled receptor 158 |
| chr14_-_55204023 | 2.05 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr14_-_55204383 | 2.04 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr9_+_54606798 | 2.03 |
ENSMUST00000154690.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr13_+_55517545 | 2.03 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr7_-_140402037 | 2.01 |
ENSMUST00000106052.2
ENSMUST00000080651.13 |
Zfp941
|
zinc finger protein 941 |
| chr14_-_79718890 | 2.00 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr14_+_26959975 | 1.89 |
ENSMUST00000049206.6
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr7_-_30738471 | 1.88 |
ENSMUST00000162250.8
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr5_-_31359559 | 1.88 |
ENSMUST00000202929.2
ENSMUST00000201231.2 ENSMUST00000114590.8 |
Zfp513
|
zinc finger protein 513 |
| chr7_+_3439144 | 1.85 |
ENSMUST00000182222.8
|
Cacng8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr9_+_58395850 | 1.83 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr5_+_137551790 | 1.83 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr3_-_92594516 | 1.82 |
ENSMUST00000029524.4
|
Lce1d
|
late cornified envelope 1D |
| chr19_-_46561532 | 1.80 |
ENSMUST00000026009.10
ENSMUST00000236255.2 |
Arl3
|
ADP-ribosylation factor-like 3 |
| chr4_-_140780972 | 1.78 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr4_-_140781015 | 1.76 |
ENSMUST00000102491.10
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr8_-_106434565 | 1.75 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chrX_+_49930311 | 1.72 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr17_+_25946644 | 1.69 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr11_-_72252663 | 1.65 |
ENSMUST00000108503.9
|
Tekt1
|
tektin 1 |
| chr4_+_124880223 | 1.64 |
ENSMUST00000030687.8
|
Rspo1
|
R-spondin 1 |
| chr3_-_100396635 | 1.62 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr9_-_21982637 | 1.57 |
ENSMUST00000179605.9
ENSMUST00000043922.7 |
Zfp653
|
zinc finger protein 653 |
| chr4_-_87724512 | 1.56 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr10_+_21469039 | 1.56 |
ENSMUST00000057341.6
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr14_+_55131568 | 1.56 |
ENSMUST00000116476.9
ENSMUST00000022808.14 ENSMUST00000150975.8 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr18_+_36098090 | 1.53 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr4_-_87724533 | 1.52 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_3939924 | 1.50 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr15_+_102234825 | 1.49 |
ENSMUST00000165671.8
ENSMUST00000165717.4 |
Pfdn5
|
prefoldin 5 |
| chr11_+_114656189 | 1.48 |
ENSMUST00000138804.8
ENSMUST00000084368.12 |
Kif19a
|
kinesin family member 19A |
| chr15_-_99355623 | 1.47 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr4_+_107659361 | 1.47 |
ENSMUST00000106731.4
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr5_+_135835713 | 1.45 |
ENSMUST00000126232.8
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr10_-_116808514 | 1.44 |
ENSMUST00000092165.5
|
Gm10271
|
predicted gene 10271 |
| chr15_-_73114855 | 1.43 |
ENSMUST00000227686.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr19_-_5507532 | 1.43 |
ENSMUST00000236881.2
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr11_-_102837514 | 1.43 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr7_-_24145107 | 1.42 |
ENSMUST00000205776.2
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr7_+_28488380 | 1.41 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr11_-_113956996 | 1.41 |
ENSMUST00000041627.14
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr6_+_124974009 | 1.40 |
ENSMUST00000159391.8
|
Pianp
|
PILR alpha associated neural protein |
| chr1_-_78488795 | 1.38 |
ENSMUST00000170511.3
|
BC035947
|
cDNA sequence BC035947 |
| chr3_-_95214443 | 1.37 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr16_-_18052937 | 1.37 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr13_+_12580772 | 1.36 |
ENSMUST00000220811.2
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr15_+_102234551 | 1.34 |
ENSMUST00000166658.8
ENSMUST00000062492.11 |
Pfdn5
|
prefoldin 5 |
| chr7_+_105053775 | 1.34 |
ENSMUST00000033187.6
ENSMUST00000210344.2 |
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr13_-_59879548 | 1.34 |
ENSMUST00000052978.6
|
Spata31d1d
|
spermatogenesis associated 31 subfamily D, member 1D |
| chr1_+_133965228 | 1.32 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr13_-_26954110 | 1.30 |
ENSMUST00000055915.6
|
Hdgfl1
|
HDGF like 1 |
| chr4_-_42665763 | 1.29 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr7_-_24134919 | 1.29 |
ENSMUST00000080594.8
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr4_-_42168603 | 1.27 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chrX_+_47001264 | 1.26 |
ENSMUST00000001202.15
ENSMUST00000115020.8 |
Ocrl
|
OCRL, inositol polyphosphate-5-phosphatase |
| chr18_+_37580692 | 1.22 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr14_-_52258158 | 1.22 |
ENSMUST00000228580.2
ENSMUST00000226554.2 ENSMUST00000067549.15 |
Zfp219
|
zinc finger protein 219 |
| chr11_-_51153767 | 1.21 |
ENSMUST00000065950.10
|
BC049762
|
cDNA sequence BC049762 |
| chr3_-_95799242 | 1.21 |
ENSMUST00000171519.2
ENSMUST00000036360.13 ENSMUST00000090476.10 |
BC028528
|
cDNA sequence BC028528 |
| chr2_+_144441817 | 1.21 |
ENSMUST00000028917.7
|
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr7_-_44785709 | 1.18 |
ENSMUST00000211429.2
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr12_+_111373243 | 1.17 |
ENSMUST00000150384.3
|
Lbhd2
|
LBH domain containing 2 |
| chr17_-_56607250 | 1.17 |
ENSMUST00000233911.2
|
Arrdc5
|
arrestin domain containing 5 |
| chr2_-_164670452 | 1.16 |
ENSMUST00000017911.4
|
Spata25
|
spermatogenesis associated 25 |
| chr4_+_137720326 | 1.14 |
ENSMUST00000139759.8
ENSMUST00000058133.10 ENSMUST00000105830.9 ENSMUST00000084215.12 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr16_-_44979013 | 1.13 |
ENSMUST00000023344.10
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr8_+_94899292 | 1.13 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr14_+_56905698 | 1.12 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr16_-_38370535 | 1.12 |
ENSMUST00000036210.7
|
Poglut1
|
protein O-glucosyltransferase 1 |
| chr18_+_37575553 | 1.10 |
ENSMUST00000056915.3
|
Pcdhb13
|
protocadherin beta 13 |
| chr9_+_22322875 | 1.10 |
ENSMUST00000214436.2
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr15_+_102234589 | 1.09 |
ENSMUST00000170627.8
|
Pfdn5
|
prefoldin 5 |
| chr6_+_113600920 | 1.09 |
ENSMUST00000035673.8
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr4_+_117692583 | 1.08 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_+_99023775 | 1.06 |
ENSMUST00000230628.2
ENSMUST00000229073.2 ENSMUST00000229147.2 |
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr18_+_37488174 | 1.05 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr15_+_58004793 | 1.03 |
ENSMUST00000227142.2
ENSMUST00000226955.2 |
Wdyhv1
|
WDYHV motif containing 1 |
| chr5_-_72325482 | 1.01 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chr17_+_35069347 | 1.01 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr7_+_140427729 | 1.00 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr6_+_113508636 | 1.00 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr10_+_70080913 | 0.99 |
ENSMUST00000046807.7
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr16_-_56748424 | 0.98 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr2_+_52747855 | 0.97 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr11_-_51153870 | 0.97 |
ENSMUST00000054226.3
|
BC049762
|
cDNA sequence BC049762 |
| chr6_-_112466780 | 0.96 |
ENSMUST00000053306.8
|
Oxtr
|
oxytocin receptor |
| chr8_-_13940234 | 0.94 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr15_+_103411689 | 0.94 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr7_+_140427711 | 0.93 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr13_+_12580743 | 0.92 |
ENSMUST00000221560.2
ENSMUST00000071973.8 |
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr15_+_58004753 | 0.90 |
ENSMUST00000067563.9
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr18_-_15196612 | 0.88 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chrX_-_151820545 | 0.88 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr15_-_48655329 | 0.87 |
ENSMUST00000160658.8
ENSMUST00000100670.10 ENSMUST00000162830.8 |
Csmd3
|
CUB and Sushi multiple domains 3 |
| chr10_+_19588318 | 0.87 |
ENSMUST00000020185.5
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr10_-_128505096 | 0.87 |
ENSMUST00000238610.2
ENSMUST00000238712.2 |
Ikzf4
|
IKAROS family zinc finger 4 |
| chr14_+_55132061 | 0.86 |
ENSMUST00000172557.2
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr9_+_66257747 | 0.83 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr7_-_44785815 | 0.83 |
ENSMUST00000146760.7
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr12_-_69629758 | 0.83 |
ENSMUST00000058639.11
|
Vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr9_+_108660989 | 0.82 |
ENSMUST00000192307.6
ENSMUST00000193560.6 ENSMUST00000194875.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr11_+_76563281 | 0.81 |
ENSMUST00000056184.2
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr16_-_44978816 | 0.79 |
ENSMUST00000181750.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr15_+_102234802 | 0.79 |
ENSMUST00000169637.8
|
Pfdn5
|
prefoldin 5 |
| chr4_+_136013372 | 0.79 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr11_-_83320281 | 0.78 |
ENSMUST00000052521.9
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr14_+_55132030 | 0.78 |
ENSMUST00000141446.8
ENSMUST00000139985.8 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr13_-_98773862 | 0.78 |
ENSMUST00000050389.5
|
Tmem174
|
transmembrane protein 174 |
| chr7_-_103792462 | 0.76 |
ENSMUST00000057254.6
|
Ubqln3
|
ubiquilin 3 |
| chr1_-_135358228 | 0.76 |
ENSMUST00000041023.14
|
Ipo9
|
importin 9 |
| chr16_-_44978929 | 0.74 |
ENSMUST00000181177.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr3_-_129548954 | 0.72 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr6_-_139478919 | 0.72 |
ENSMUST00000170650.3
|
Rergl
|
RERG/RAS-like |
| chr19_-_23251179 | 0.72 |
ENSMUST00000087556.7
ENSMUST00000223934.2 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr8_-_95564881 | 0.71 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr17_-_25652750 | 0.71 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr11_-_17903861 | 0.71 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr11_+_51153941 | 0.70 |
ENSMUST00000093132.13
ENSMUST00000109113.8 |
Clk4
|
CDC like kinase 4 |
| chr3_-_107424637 | 0.70 |
ENSMUST00000166892.2
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr18_+_37646674 | 0.69 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr2_+_32340459 | 0.68 |
ENSMUST00000048431.3
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr17_-_56607286 | 0.66 |
ENSMUST00000097303.3
|
Arrdc5
|
arrestin domain containing 5 |
| chr15_+_99024394 | 0.66 |
ENSMUST00000063517.6
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr10_-_120815232 | 0.64 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chrX_-_52328963 | 0.60 |
ENSMUST00000074861.9
|
Plac1
|
placental specific protein 1 |
| chr16_-_44978986 | 0.59 |
ENSMUST00000180636.8
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr7_+_79939747 | 0.58 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr8_+_120339440 | 0.58 |
ENSMUST00000098361.4
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr17_+_29171386 | 0.58 |
ENSMUST00000118762.9
ENSMUST00000057174.16 ENSMUST00000232874.2 ENSMUST00000232772.2 ENSMUST00000233334.2 ENSMUST00000150858.2 ENSMUST00000233064.2 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 2.3 | 9.1 | GO:0007522 | visceral muscle development(GO:0007522) |
| 2.1 | 8.2 | GO:1990768 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 1.9 | 5.8 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 1.5 | 9.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.5 | 9.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.3 | 6.6 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 1.3 | 3.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.2 | 8.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.9 | 5.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.9 | 4.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.8 | 3.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.7 | 2.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.6 | 2.5 | GO:2001107 | negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.6 | 3.5 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 2.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.4 | 8.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.4 | 7.1 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.4 | 6.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.4 | 1.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.4 | 1.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 2.7 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.3 | 2.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 4.3 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.3 | 7.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 1.4 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.3 | 3.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 0.7 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 | 2.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 2.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 4.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 3.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 1.7 | GO:0031119 | rRNA pseudouridine synthesis(GO:0031118) tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 2.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.2 | 6.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 1.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.6 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 2.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.7 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.6 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 2.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.9 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 3.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 3.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.4 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 1.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.3 | GO:0070318 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 2.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.5 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.6 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 2.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 1.0 | GO:1900364 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 2.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.6 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 4.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.6 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.7 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 0.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 2.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 4.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.7 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 3.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 2.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 1.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 4.8 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 7.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 7.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 5.0 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 2.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 4.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 5.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 1.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.9 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.7 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 1.2 | GO:0048477 | oogenesis(GO:0048477) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 9.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.1 | 9.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.9 | 2.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.7 | 2.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.4 | 4.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 6.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 9.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 1.4 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.3 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 5.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 1.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 4.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 1.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 4.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 3.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 5.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 3.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 3.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 3.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 5.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 3.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 4.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 7.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 11.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 6.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 7.7 | GO:0005912 | adherens junction(GO:0005912) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 1.5 | 9.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.5 | 5.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 1.3 | 9.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.8 | 9.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.7 | 2.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.7 | 2.2 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.6 | 5.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 8.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 2.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.6 | 7.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.5 | 4.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 3.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 4.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 3.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.4 | 1.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 1.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.4 | 1.5 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 3.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 0.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 4.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.3 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 2.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 1.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 3.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.6 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.8 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 2.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 2.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 3.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.4 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.1 | 6.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 14.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 3.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 3.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 9.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 2.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 9.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 4.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 5.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 2.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 9.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 4.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 9.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 3.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 4.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 4.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 9.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 6.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 3.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 5.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |