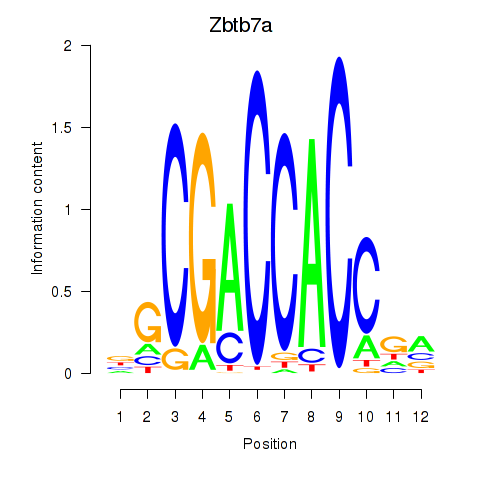
Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zbtb7a
Z-value: 1.61
Transcription factors associated with Zbtb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7a
|
ENSMUSG00000035011.16 | Zbtb7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7a | mm39_v1_chr10_+_80972089_80972105 | 0.46 | 4.7e-05 | Click! |
Activity profile of Zbtb7a motif
Sorted Z-values of Zbtb7a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_127098932 | 16.20 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr10_-_127099183 | 16.14 |
ENSMUST00000099172.5
|
Kif5a
|
kinesin family member 5A |
| chr11_-_70924288 | 12.47 |
ENSMUST00000238695.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr2_+_157401998 | 10.31 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr9_+_45281483 | 9.43 |
ENSMUST00000085939.8
ENSMUST00000217381.2 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr9_+_102988940 | 9.01 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr2_-_163760603 | 8.99 |
ENSMUST00000044734.3
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr1_-_38875757 | 8.54 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr2_-_155356716 | 8.51 |
ENSMUST00000029131.11
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr1_-_75240551 | 8.41 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr11_+_101066867 | 8.38 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr7_-_30826184 | 8.31 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr11_+_53410552 | 8.29 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr12_+_44375747 | 7.81 |
ENSMUST00000020939.16
ENSMUST00000220126.2 |
Nrcam
|
neuronal cell adhesion molecule |
| chr7_-_57159119 | 7.58 |
ENSMUST00000206382.2
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr12_-_4088905 | 7.55 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chr9_+_86454018 | 7.54 |
ENSMUST00000185566.7
ENSMUST00000034988.10 ENSMUST00000179212.3 |
Rwdd2a
|
RWD domain containing 2A |
| chr1_+_87192067 | 7.54 |
ENSMUST00000027472.7
|
Efhd1
|
EF hand domain containing 1 |
| chr12_-_109034099 | 7.47 |
ENSMUST00000190647.3
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr8_+_120173458 | 7.23 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr12_+_108300599 | 7.22 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr19_-_5560473 | 7.11 |
ENSMUST00000237111.2
ENSMUST00000070172.6 |
Snx32
|
sorting nexin 32 |
| chrX_+_165021919 | 7.03 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr7_-_81584081 | 6.95 |
ENSMUST00000026094.6
ENSMUST00000107305.8 |
Hdgfl3
|
HDGF like 3 |
| chr18_+_33072194 | 6.92 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr4_-_155482516 | 6.61 |
ENSMUST00000030925.3
|
Gabrd
|
gamma-aminobutyric acid (GABA) A receptor, subunit delta |
| chr11_+_53410697 | 6.55 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr18_+_63841756 | 6.52 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr11_+_68582731 | 6.39 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr11_+_68582923 | 6.36 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr2_+_150628655 | 6.21 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr11_-_74480870 | 6.04 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr12_+_44375665 | 6.03 |
ENSMUST00000110748.4
|
Nrcam
|
neuronal cell adhesion molecule |
| chr12_+_51640097 | 5.94 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr7_-_57159743 | 5.92 |
ENSMUST00000068456.8
ENSMUST00000206734.2 |
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr2_-_25209107 | 5.91 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr13_-_99653045 | 5.86 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr7_+_44091822 | 5.81 |
ENSMUST00000058667.15
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr6_-_144994534 | 5.77 |
ENSMUST00000032402.12
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr8_+_123844090 | 5.74 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chr15_+_89407954 | 5.70 |
ENSMUST00000230807.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chrX_+_165021897 | 5.65 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr4_-_155859014 | 5.62 |
ENSMUST00000042196.4
|
Vwa1
|
von Willebrand factor A domain containing 1 |
| chr19_-_5135510 | 5.55 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr19_+_44282113 | 5.49 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr15_+_78783867 | 5.46 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr19_-_40600619 | 5.30 |
ENSMUST00000132452.2
ENSMUST00000135795.8 ENSMUST00000025981.15 |
Tctn3
|
tectonic family member 3 |
| chr10_-_76073656 | 5.29 |
ENSMUST00000099572.10
ENSMUST00000020452.12 ENSMUST00000099571.10 |
Prmt2
|
protein arginine N-methyltransferase 2 |
| chr11_+_68582897 | 5.24 |
ENSMUST00000108673.8
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr1_-_135302971 | 5.21 |
ENSMUST00000041240.4
|
Shisa4
|
shisa family member 4 |
| chr5_+_37403098 | 5.15 |
ENSMUST00000031004.11
|
Crmp1
|
collapsin response mediator protein 1 |
| chr14_+_111912529 | 5.14 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr17_+_6320731 | 5.09 |
ENSMUST00000088940.6
|
Tmem181a
|
transmembrane protein 181A |
| chr7_-_112968533 | 5.05 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr4_+_42917228 | 5.04 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr4_-_58553311 | 5.01 |
ENSMUST00000107571.8
ENSMUST00000055018.11 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr18_+_36098090 | 4.92 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr3_+_123240562 | 4.89 |
ENSMUST00000029603.10
|
Prss12
|
protease, serine 12 neurotrypsin (motopsin) |
| chr19_-_42420216 | 4.87 |
ENSMUST00000048630.8
ENSMUST00000238290.2 |
Crtac1
|
cartilage acidic protein 1 |
| chr18_+_86729645 | 4.82 |
ENSMUST00000122464.8
|
Cbln2
|
cerebellin 2 precursor protein |
| chr17_+_44112187 | 4.82 |
ENSMUST00000228972.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr19_+_47217279 | 4.80 |
ENSMUST00000111807.5
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr6_+_125192514 | 4.76 |
ENSMUST00000032487.14
ENSMUST00000100942.9 ENSMUST00000063588.11 |
Vamp1
|
vesicle-associated membrane protein 1 |
| chr11_+_120612369 | 4.75 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr6_-_42669963 | 4.73 |
ENSMUST00000045140.5
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr18_+_4634878 | 4.66 |
ENSMUST00000037029.7
|
Jcad
|
junctional cadherin 5 associated |
| chr6_-_144994327 | 4.62 |
ENSMUST00000204138.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr3_-_32670628 | 4.61 |
ENSMUST00000193050.2
ENSMUST00000108234.8 ENSMUST00000155737.8 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr17_+_27904155 | 4.58 |
ENSMUST00000231669.2
ENSMUST00000097360.3 ENSMUST00000231236.2 |
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr11_+_120612278 | 4.55 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr14_-_55231998 | 4.55 |
ENSMUST00000227518.2
ENSMUST00000226424.2 ENSMUST00000153783.2 ENSMUST00000102803.11 ENSMUST00000168485.8 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr7_-_30826376 | 4.51 |
ENSMUST00000098548.8
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr5_-_71815318 | 4.50 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr1_-_170417354 | 4.49 |
ENSMUST00000160456.8
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr5_+_142946598 | 4.48 |
ENSMUST00000129306.4
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr5_-_18565353 | 4.48 |
ENSMUST00000074694.7
|
Gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
| chr1_+_172139934 | 4.47 |
ENSMUST00000039506.15
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr17_-_24908874 | 4.44 |
ENSMUST00000007236.5
|
Syngr3
|
synaptogyrin 3 |
| chr4_-_58553553 | 4.44 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr9_-_108455899 | 4.40 |
ENSMUST00000068700.7
|
Wdr6
|
WD repeat domain 6 |
| chr4_+_120711974 | 4.37 |
ENSMUST00000071093.9
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr8_-_106198112 | 4.34 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_-_99450693 | 4.32 |
ENSMUST00000185524.7
ENSMUST00000186049.2 |
Armc8
|
armadillo repeat containing 8 |
| chr7_+_126528016 | 4.27 |
ENSMUST00000032924.6
|
Kctd13
|
potassium channel tetramerisation domain containing 13 |
| chr19_+_40600836 | 4.26 |
ENSMUST00000134063.8
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr7_+_15864265 | 4.21 |
ENSMUST00000168693.3
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr16_+_91066602 | 4.19 |
ENSMUST00000056882.7
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr8_-_26484131 | 4.16 |
ENSMUST00000061850.5
|
Pomk
|
protein-O-mannose kinase |
| chr13_-_49301407 | 4.15 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr11_-_121279062 | 4.14 |
ENSMUST00000106107.3
|
Rab40b
|
Rab40B, member RAS oncogene family |
| chr8_-_13939964 | 4.13 |
ENSMUST00000209371.2
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr18_+_86729184 | 4.12 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr4_-_151192911 | 4.10 |
ENSMUST00000105670.8
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr11_-_35689711 | 4.09 |
ENSMUST00000160726.4
|
Fbll1
|
fibrillarin-like 1 |
| chr6_+_4747298 | 4.07 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chr7_+_40547608 | 4.04 |
ENSMUST00000044705.12
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr19_+_4149998 | 4.00 |
ENSMUST00000049658.14
|
Pitpnm1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr9_+_106080307 | 4.00 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr4_-_150039486 | 3.97 |
ENSMUST00000038562.9
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr7_+_141503411 | 3.93 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr7_-_27628855 | 3.87 |
ENSMUST00000098639.9
|
Zfp974
|
zinc finger protein 974 |
| chrX_+_98086187 | 3.86 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr1_+_17797257 | 3.79 |
ENSMUST00000159958.8
ENSMUST00000160305.8 ENSMUST00000095075.5 |
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr15_-_84441977 | 3.76 |
ENSMUST00000069476.5
|
Rtl6
|
retrotransposon Gag like 6 |
| chr6_+_135175031 | 3.67 |
ENSMUST00000130612.2
|
Fam234b
|
family with sequence similarity 234, member B |
| chr7_+_5054514 | 3.64 |
ENSMUST00000069324.7
|
Zfp580
|
zinc finger protein 580 |
| chr15_+_100768806 | 3.64 |
ENSMUST00000201549.4
ENSMUST00000108908.6 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr5_+_149335214 | 3.62 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr16_+_17379749 | 3.55 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr3_+_87855973 | 3.50 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr3_-_51303924 | 3.50 |
ENSMUST00000108046.8
ENSMUST00000038154.12 |
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr7_-_109559593 | 3.49 |
ENSMUST00000106722.2
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr14_-_34096574 | 3.49 |
ENSMUST00000023826.5
|
Sncg
|
synuclein, gamma |
| chr13_-_70785753 | 3.45 |
ENSMUST00000043493.7
|
Ice1
|
interactor of little elongation complex ELL subunit 1 |
| chr2_-_32271833 | 3.44 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr19_-_4333061 | 3.42 |
ENSMUST00000167215.2
ENSMUST00000056888.13 |
Ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr14_-_51022952 | 3.41 |
ENSMUST00000226768.2
ENSMUST00000006451.8 |
Ttc5
|
tetratricopeptide repeat domain 5 |
| chr7_-_81104423 | 3.40 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr13_+_54722823 | 3.36 |
ENSMUST00000026988.11
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr4_-_128856213 | 3.35 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr2_-_118203826 | 3.35 |
ENSMUST00000039160.3
|
Gpr176
|
G protein-coupled receptor 176 |
| chr19_-_5323092 | 3.35 |
ENSMUST00000237463.2
ENSMUST00000025786.9 |
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr8_-_69187708 | 3.32 |
ENSMUST00000136060.8
ENSMUST00000130214.8 ENSMUST00000078350.13 |
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_13940234 | 3.32 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr5_+_118165808 | 3.31 |
ENSMUST00000031304.14
|
Tesc
|
tescalcin |
| chr9_+_67747668 | 3.29 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chr6_+_4747356 | 3.28 |
ENSMUST00000176551.3
|
Peg10
|
paternally expressed 10 |
| chr4_+_103476777 | 3.27 |
ENSMUST00000106827.8
|
Dab1
|
disabled 1 |
| chr6_+_135174975 | 3.24 |
ENSMUST00000111915.8
ENSMUST00000111916.2 |
Fam234b
|
family with sequence similarity 234, member B |
| chr13_+_54722833 | 3.24 |
ENSMUST00000156024.2
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr12_-_111638722 | 3.22 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chrX_-_149596680 | 3.17 |
ENSMUST00000112700.8
|
Maged2
|
MAGE family member D2 |
| chr6_-_143045731 | 3.16 |
ENSMUST00000203673.3
ENSMUST00000203187.3 ENSMUST00000171349.8 ENSMUST00000087485.7 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr3_+_105359641 | 3.15 |
ENSMUST00000098761.10
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr2_+_24944407 | 3.15 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr19_-_61215743 | 3.13 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr10_-_57408512 | 3.13 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr11_+_104122399 | 3.13 |
ENSMUST00000132977.8
ENSMUST00000132245.8 ENSMUST00000100347.11 |
Mapt
|
microtubule-associated protein tau |
| chr15_-_76702170 | 3.13 |
ENSMUST00000175843.3
ENSMUST00000177026.3 ENSMUST00000176736.3 ENSMUST00000036176.16 ENSMUST00000176219.9 ENSMUST00000239134.2 ENSMUST00000239003.2 ENSMUST00000077821.10 |
Arhgap39
|
Rho GTPase activating protein 39 |
| chr13_-_95359543 | 3.11 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr15_-_79658608 | 3.11 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr8_+_33876353 | 3.09 |
ENSMUST00000070340.6
ENSMUST00000078058.5 |
Purg
|
purine-rich element binding protein G |
| chr17_+_87415049 | 3.08 |
ENSMUST00000041369.8
ENSMUST00000234803.2 |
Socs5
|
suppressor of cytokine signaling 5 |
| chr2_+_26205525 | 3.08 |
ENSMUST00000066936.9
ENSMUST00000078616.12 |
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr15_+_32244947 | 3.04 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr11_+_3282424 | 3.04 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_-_181333597 | 3.03 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr2_-_105229653 | 3.01 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr19_+_23736205 | 3.00 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr7_-_6699422 | 2.99 |
ENSMUST00000122432.4
ENSMUST00000002336.16 |
Zim1
|
zinc finger, imprinted 1 |
| chr16_+_75389732 | 2.96 |
ENSMUST00000046378.14
ENSMUST00000114249.8 ENSMUST00000114253.2 |
Rbm11
|
RNA binding motif protein 11 |
| chr11_+_104122341 | 2.92 |
ENSMUST00000106993.10
|
Mapt
|
microtubule-associated protein tau |
| chr12_-_110807330 | 2.92 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr10_-_79710067 | 2.92 |
ENSMUST00000166023.2
ENSMUST00000167707.2 ENSMUST00000165601.8 |
Plppr3
|
phospholipid phosphatase related 3 |
| chr13_-_14697770 | 2.91 |
ENSMUST00000110516.3
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr11_-_70578775 | 2.89 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chrX_+_55391749 | 2.88 |
ENSMUST00000101560.4
|
Zfp449
|
zinc finger protein 449 |
| chr1_+_128031055 | 2.86 |
ENSMUST00000188381.7
ENSMUST00000187900.7 ENSMUST00000036288.11 |
R3hdm1
|
R3H domain containing 1 |
| chr12_-_76756772 | 2.85 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr6_-_4747157 | 2.85 |
ENSMUST00000126151.8
ENSMUST00000115577.9 ENSMUST00000101677.9 ENSMUST00000115579.8 ENSMUST00000004750.15 |
Sgce
|
sarcoglycan, epsilon |
| chr4_+_152171286 | 2.84 |
ENSMUST00000118648.8
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr12_-_28800363 | 2.81 |
ENSMUST00000170994.3
|
Trappc12
|
trafficking protein particle complex 12 |
| chr11_+_117545037 | 2.80 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr7_-_141925947 | 2.80 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr12_-_28800424 | 2.80 |
ENSMUST00000020954.15
ENSMUST00000168129.10 |
Trappc12
|
trafficking protein particle complex 12 |
| chr13_-_78347876 | 2.79 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr5_-_34445662 | 2.78 |
ENSMUST00000094868.10
|
Zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr15_+_100768776 | 2.78 |
ENSMUST00000108909.9
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr11_-_97877219 | 2.78 |
ENSMUST00000107565.3
ENSMUST00000107564.2 ENSMUST00000017561.15 |
Plxdc1
|
plexin domain containing 1 |
| chr15_+_99122742 | 2.78 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr11_+_78079631 | 2.78 |
ENSMUST00000056241.12
ENSMUST00000207728.2 |
Rab34
|
RAB34, member RAS oncogene family |
| chr10_-_49664839 | 2.76 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr15_-_79658584 | 2.76 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_-_25209199 | 2.76 |
ENSMUST00000114312.2
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr11_-_100331884 | 2.74 |
ENSMUST00000107399.9
ENSMUST00000092688.12 |
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr9_+_66257747 | 2.73 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr6_+_83055581 | 2.73 |
ENSMUST00000177177.8
ENSMUST00000176089.2 |
Pcgf1
|
polycomb group ring finger 1 |
| chr6_+_85164420 | 2.71 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr9_+_40180726 | 2.68 |
ENSMUST00000171835.9
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr12_-_85224062 | 2.67 |
ENSMUST00000004913.7
|
Pgf
|
placental growth factor |
| chr11_-_33942981 | 2.65 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr6_-_124888643 | 2.64 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr4_+_127062924 | 2.63 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr11_+_84848601 | 2.61 |
ENSMUST00000103194.10
|
Car4
|
carbonic anhydrase 4 |
| chr2_+_24944367 | 2.60 |
ENSMUST00000100334.11
ENSMUST00000152122.8 ENSMUST00000116574.10 ENSMUST00000006646.15 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chrX_-_149595711 | 2.58 |
ENSMUST00000112697.10
|
Maged2
|
MAGE family member D2 |
| chr11_-_100331754 | 2.57 |
ENSMUST00000107397.2
ENSMUST00000092689.9 |
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr7_+_18810167 | 2.54 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr4_-_138802868 | 2.54 |
ENSMUST00000105802.8
|
Htr6
|
5-hydroxytryptamine (serotonin) receptor 6 |
| chr11_-_120534469 | 2.49 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr9_+_31191820 | 2.49 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr7_+_6289572 | 2.49 |
ENSMUST00000086327.12
ENSMUST00000153840.2 ENSMUST00000170776.8 |
Zfp667
|
zinc finger protein 667 |
| chr9_-_108067552 | 2.49 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr2_-_29142965 | 2.48 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr9_+_124195807 | 2.48 |
ENSMUST00000239563.2
|
Ppp2r3d
|
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
| chr12_-_98867737 | 2.45 |
ENSMUST00000223282.2
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr5_-_108515740 | 2.44 |
ENSMUST00000197216.3
|
Gm42517
|
predicted gene 42517 |
| chr5_+_124736812 | 2.42 |
ENSMUST00000239501.2
|
Tctn2
|
tectonic family member 2 |
| chr4_+_136011969 | 2.41 |
ENSMUST00000144217.8
|
Zfp46
|
zinc finger protein 46 |
| chr10_-_57408585 | 2.41 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.3 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 4.5 | 18.0 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 4.3 | 12.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 2.9 | 8.7 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 2.4 | 7.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 2.4 | 9.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.6 | 12.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.5 | 4.6 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 1.5 | 18.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.5 | 4.5 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 1.4 | 4.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 1.3 | 10.4 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 1.3 | 5.1 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.2 | 3.5 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 1.1 | 3.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 1.1 | 3.3 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.1 | 7.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 1.0 | 8.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.0 | 4.9 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 1.0 | 5.7 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.9 | 2.8 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.9 | 5.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.8 | 9.1 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.8 | 2.5 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.8 | 3.3 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.8 | 5.7 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.8 | 5.7 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.8 | 8.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.8 | 5.6 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.8 | 8.5 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.7 | 0.7 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.7 | 4.3 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.7 | 2.8 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.7 | 2.0 | GO:0046959 | habituation(GO:0046959) |
| 0.7 | 4.7 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.7 | 4.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.7 | 5.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.6 | 4.5 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.6 | 4.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.6 | 5.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.6 | 8.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.6 | 4.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 2.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.6 | 1.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 1.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.6 | 2.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.6 | 2.2 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.6 | 3.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.5 | 4.4 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.5 | 8.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.5 | 1.0 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.5 | 2.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.5 | 3.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.5 | 6.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.5 | 2.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 4.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.5 | 4.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.5 | 2.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.5 | 2.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 7.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 3.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.4 | 2.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.4 | 5.3 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.4 | 3.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 2.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.4 | 17.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 4.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.4 | 2.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 1.2 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.4 | 2.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 2.3 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.4 | 3.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.4 | 1.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.7 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.3 | 2.4 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.3 | 0.7 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.3 | 14.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 2.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.3 | 2.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 1.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 5.9 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.3 | 5.1 | GO:0021756 | striatum development(GO:0021756) |
| 0.3 | 2.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 13.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 3.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 1.5 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 2.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 3.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 6.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.3 | 2.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.3 | 7.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 7.8 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.3 | 4.5 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.3 | 3.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 3.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 2.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.3 | 6.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 0.8 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.3 | 1.9 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.3 | 3.2 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.3 | 0.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.2 | 3.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 2.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 7.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 2.8 | GO:0090500 | dorsal aorta morphogenesis(GO:0035912) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.2 | 3.2 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.2 | 1.6 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 2.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 1.1 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.2 | 7.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 2.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.2 | 0.8 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 8.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 2.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.2 | 3.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 1.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 4.8 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 0.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 1.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 1.9 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 4.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.2 | 3.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 | 4.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.2 | 1.2 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.2 | 6.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 3.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 | 2.7 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 0.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 7.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.5 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 1.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 5.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 12.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 2.3 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.8 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 2.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 6.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 14.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 3.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 10.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 3.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 5.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 3.3 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.1 | 2.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.4 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.1 | 1.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.8 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 2.1 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 5.2 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.1 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 2.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 1.0 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) serotonin metabolic process(GO:0042428) |
| 0.1 | 1.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 2.6 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 4.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.1 | 3.0 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 3.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.8 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 3.5 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 4.3 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 1.6 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 1.1 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 4.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.2 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 2.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 4.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 7.9 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.0 | 1.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.9 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.1 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0051771 | protein geranylgeranylation(GO:0018344) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 2.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 2.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.3 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 4.7 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 2.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 3.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 2.5 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.0 | 0.2 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 1.1 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 1.6 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.0 | 4.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 6.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.1 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) telomerase RNA stabilization(GO:0090669) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 2.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.5 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.7 | 8.7 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.2 | 31.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.0 | 3.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 1.0 | 26.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.9 | 4.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.9 | 4.5 | GO:0044393 | microspike(GO:0044393) |
| 0.9 | 8.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.9 | 22.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 4.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.8 | 2.5 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.8 | 3.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.7 | 3.4 | GO:0035363 | histone locus body(GO:0035363) |
| 0.5 | 4.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 3.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 2.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 1.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 4.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 8.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.4 | 16.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 1.5 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.3 | 3.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 2.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 2.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.3 | 4.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 1.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 5.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 4.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 2.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.3 | 0.8 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.3 | 5.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 0.7 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.2 | 4.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 5.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.7 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 14.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 6.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 3.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 2.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 10.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 13.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 0.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 2.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 2.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 2.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 38.3 | GO:0044309 | neuron spine(GO:0044309) |
| 0.1 | 2.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 4.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 4.0 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 4.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 8.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.9 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 2.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 6.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 3.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 14.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) germinal vesicle(GO:0042585) |
| 0.1 | 4.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 18.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 4.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 11.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 4.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 4.1 | GO:0030135 | coated vesicle(GO:0030135) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 9.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 13.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 4.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 3.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 2.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.8 | 7.2 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 1.6 | 8.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.6 | 6.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.4 | 41.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.4 | 4.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 1.3 | 9.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.3 | 4.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 1.3 | 10.4 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.3 | 13.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.2 | 4.8 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 1.1 | 8.7 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.0 | 3.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.9 | 5.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.9 | 5.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.9 | 6.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.8 | 23.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.8 | 3.3 | GO:0047238 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.8 | 21.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.8 | 5.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.8 | 3.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.8 | 5.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 4.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 8.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.7 | 2.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.7 | 2.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.7 | 5.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.6 | 3.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.6 | 1.8 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.6 | 1.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.6 | 1.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.6 | 2.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.5 | 2.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 4.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 2.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 3.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 4.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.5 | 4.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.5 | 2.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 2.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 4.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 5.7 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 6.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 2.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 1.6 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 2.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 3.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 1.5 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.4 | 2.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 4.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 5.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 2.4 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.3 | 9.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 4.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 2.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 6.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 1.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 2.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.3 | 1.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.3 | 2.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 0.9 | GO:0070698 | nodal binding(GO:0038100) type I activin receptor binding(GO:0070698) |
| 0.3 | 5.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 0.8 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.3 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 7.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.2 | 1.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 5.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 0.6 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 4.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 5.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 3.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 2.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 4.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 6.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 7.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 2.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 9.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.8 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 4.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 10.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.7 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 8.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 15.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.5 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 7.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 4.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 3.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 23.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 4.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 3.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 13.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 2.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 2.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 4.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 5.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 11.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 4.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 4.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 1.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 14.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 9.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 10.0 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 1.6 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 4.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 3.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 21.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 2.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 5.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 5.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 9.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 13.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 8.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 13.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 3.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 3.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 13.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 6.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 6.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 32.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.8 | 20.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 39.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 18.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.5 | 7.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 5.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 8.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 6.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 7.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 8.8 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.3 | 4.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 3.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 3.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 2.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 4.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 2.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 5.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 6.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 6.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 8.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 7.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 8.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 6.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 3.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.8 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 1.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 13.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 2.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 8.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 7.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |