Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zbtb7b
Z-value: 0.95
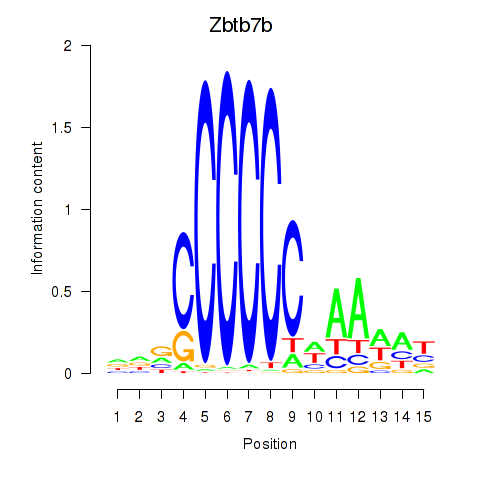
Transcription factors associated with Zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7b
|
ENSMUSG00000028042.16 | Zbtb7b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | mm39_v1_chr3_-_89300599_89300685 | -0.57 | 1.5e-07 | Click! |
Activity profile of Zbtb7b motif
Sorted Z-values of Zbtb7b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_17712061 | 8.17 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr1_-_74974707 | 8.11 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65 |
| chr15_-_74508197 | 7.92 |
ENSMUST00000023271.8
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chr3_+_31204069 | 7.32 |
ENSMUST00000046174.8
|
Cldn11
|
claudin 11 |
| chr11_-_82655132 | 7.08 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr17_+_33651864 | 6.52 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr2_-_152422220 | 6.45 |
ENSMUST00000053180.4
|
Defb19
|
defensin beta 19 |
| chr17_-_28298721 | 6.38 |
ENSMUST00000233898.2
ENSMUST00000232798.2 |
Tcp11
|
t-complex protein 11 |
| chr11_-_103109247 | 6.35 |
ENSMUST00000103076.2
|
Spata32
|
spermatogenesis associated 32 |
| chr5_-_123620632 | 6.02 |
ENSMUST00000198901.2
|
Il31
|
interleukin 31 |
| chr10_+_84591919 | 5.84 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr7_+_46636562 | 5.68 |
ENSMUST00000185832.2
|
Gm9999
|
predicted gene 9999 |
| chr8_-_106140106 | 5.52 |
ENSMUST00000167294.8
ENSMUST00000063071.13 |
Kctd19
|
potassium channel tetramerisation domain containing 19 |
| chr6_-_113694633 | 5.37 |
ENSMUST00000204533.3
|
Ghrl
|
ghrelin |
| chr15_-_89310060 | 5.06 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chrX_+_165021919 | 4.90 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr12_+_113038098 | 4.89 |
ENSMUST00000012355.14
ENSMUST00000146107.8 |
Tex22
|
testis expressed gene 22 |
| chr11_-_97944239 | 4.88 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr13_-_112788829 | 4.87 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4 |
| chr7_+_140427729 | 4.83 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr8_+_84075066 | 4.75 |
ENSMUST00000038692.6
|
Mgat4d
|
MGAT4 family, member C |
| chr19_-_8816530 | 4.60 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr12_+_113038376 | 4.56 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chrX_+_165021897 | 4.56 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr7_-_92523396 | 4.55 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr13_-_48779047 | 4.52 |
ENSMUST00000222028.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr11_-_42073737 | 4.51 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr11_+_70506674 | 4.49 |
ENSMUST00000180052.8
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr19_-_42074777 | 4.46 |
ENSMUST00000051772.10
|
Morn4
|
MORN repeat containing 4 |
| chr11_+_70506716 | 4.34 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr11_+_104077153 | 4.34 |
ENSMUST00000107000.2
ENSMUST00000059448.8 |
Sppl2c
|
signal peptide peptidase 2C |
| chr7_-_133966588 | 4.32 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr6_-_72935171 | 4.32 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr11_-_42072990 | 4.27 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr6_+_116241146 | 4.22 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr19_+_36532061 | 4.19 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr4_-_128856213 | 4.18 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr7_+_140427711 | 4.12 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr8_+_78939920 | 4.07 |
ENSMUST00000117845.8
ENSMUST00000126172.8 ENSMUST00000049395.15 |
Ttc29
|
tetratricopeptide repeat domain 29 |
| chr15_+_76152276 | 3.86 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr7_-_134100659 | 3.78 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr9_-_121668527 | 3.77 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr8_-_105036664 | 3.77 |
ENSMUST00000160596.8
ENSMUST00000164175.2 |
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr10_-_21943978 | 3.74 |
ENSMUST00000092672.6
|
4930444G20Rik
|
RIKEN cDNA 4930444G20 gene |
| chrX_+_93278526 | 3.67 |
ENSMUST00000113908.8
ENSMUST00000113916.10 |
Klhl15
|
kelch-like 15 |
| chr9_+_110643054 | 3.64 |
ENSMUST00000098345.3
|
Prss44
|
protease, serine 44 |
| chr7_-_4755971 | 3.56 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr15_-_76259126 | 3.52 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr1_-_10079325 | 3.46 |
ENSMUST00000176398.8
ENSMUST00000027049.10 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr11_+_85061922 | 3.34 |
ENSMUST00000018623.4
|
1700125H20Rik
|
RIKEN cDNA 1700125H20 gene |
| chrX_+_139243012 | 3.22 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr7_+_44830606 | 3.22 |
ENSMUST00000210086.2
|
Pth2
|
parathyroid hormone 2 |
| chr12_-_102724931 | 3.10 |
ENSMUST00000174651.2
|
Gm20604
|
predicted gene 20604 |
| chr12_+_113038397 | 3.06 |
ENSMUST00000155492.2
|
Tex22
|
testis expressed gene 22 |
| chr19_-_45800730 | 3.05 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr2_+_91757594 | 3.03 |
ENSMUST00000045537.4
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr11_-_97909134 | 2.98 |
ENSMUST00000107561.9
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr19_-_42420216 | 2.98 |
ENSMUST00000048630.8
ENSMUST00000238290.2 |
Crtac1
|
cartilage acidic protein 1 |
| chr3_+_130904000 | 2.97 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr1_+_155433858 | 2.97 |
ENSMUST00000080138.13
ENSMUST00000035560.9 ENSMUST00000097529.5 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr4_+_129941633 | 2.95 |
ENSMUST00000044565.15
ENSMUST00000132251.2 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr13_-_48779072 | 2.86 |
ENSMUST00000035824.11
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr5_+_115568638 | 2.86 |
ENSMUST00000131079.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chrX_+_23748792 | 2.82 |
ENSMUST00000116614.3
|
Tesl1
|
testin LIM domain protein like 1 |
| chr13_+_105218625 | 2.81 |
ENSMUST00000022232.6
|
Nt5el
|
5' nucleotidase, ecto-like |
| chr14_+_65835995 | 2.76 |
ENSMUST00000150897.8
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr16_+_41353360 | 2.76 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr16_-_78373510 | 2.75 |
ENSMUST00000231973.2
ENSMUST00000232528.2 ENSMUST00000114220.9 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chrX_-_100838004 | 2.75 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779 |
| chr2_-_32665342 | 2.73 |
ENSMUST00000161089.8
ENSMUST00000066478.9 ENSMUST00000161950.8 ENSMUST00000091059.12 |
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr5_+_115568002 | 2.73 |
ENSMUST00000067168.9
|
Msi1
|
musashi RNA-binding protein 1 |
| chr7_-_4525793 | 2.71 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr19_+_8816663 | 2.68 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_-_18662526 | 2.65 |
ENSMUST00000216654.2
|
Gm4922
|
predicted gene 4922 |
| chr17_+_35454833 | 2.61 |
ENSMUST00000118384.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr6_-_134864731 | 2.56 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr11_+_4936824 | 2.54 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr9_-_62895197 | 2.54 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1 |
| chrX_+_165127688 | 2.53 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr2_+_156562956 | 2.50 |
ENSMUST00000109566.9
ENSMUST00000146412.9 ENSMUST00000177013.8 ENSMUST00000171030.9 |
Dlgap4
|
DLG associated protein 4 |
| chr5_-_115332343 | 2.42 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr7_+_44830419 | 2.41 |
ENSMUST00000042754.7
|
Pth2
|
parathyroid hormone 2 |
| chr17_+_35455532 | 2.39 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_+_119393210 | 2.35 |
ENSMUST00000033218.15
ENSMUST00000106520.9 |
Rexo5
|
RNA exonuclease 5 |
| chr11_+_94881861 | 2.35 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr15_+_97259060 | 2.32 |
ENSMUST00000228521.2
ENSMUST00000226495.2 |
Pced1b
|
PC-esterase domain containing 1B |
| chr8_-_105036739 | 2.29 |
ENSMUST00000159039.2
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr2_+_156562989 | 2.29 |
ENSMUST00000000094.14
|
Dlgap4
|
DLG associated protein 4 |
| chrX_+_90523732 | 2.25 |
ENSMUST00000101396.3
|
Mageb6b2
|
MAGE family member B6B2 |
| chr16_-_17711950 | 2.25 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr6_-_52168675 | 2.23 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr11_-_113600346 | 2.21 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr3_+_88523440 | 2.21 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr9_+_110075133 | 2.14 |
ENSMUST00000199736.2
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr2_-_117173190 | 2.12 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr3_+_88523730 | 2.08 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_84817000 | 2.06 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr9_-_103182246 | 2.02 |
ENSMUST00000142540.2
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr11_-_69768875 | 2.01 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr2_-_73605387 | 1.99 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr14_+_30673334 | 1.97 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr11_+_69909659 | 1.95 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr2_-_117173428 | 1.94 |
ENSMUST00000102534.11
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr11_-_109613040 | 1.91 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr2_-_151510453 | 1.86 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr2_-_117173312 | 1.84 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr11_-_113600838 | 1.80 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr7_-_97387145 | 1.79 |
ENSMUST00000084986.8
|
Aqp11
|
aquaporin 11 |
| chr11_+_69909245 | 1.78 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr5_+_139197689 | 1.77 |
ENSMUST00000148772.8
ENSMUST00000110882.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr6_+_41515152 | 1.77 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chrX_+_8137620 | 1.73 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_8137372 | 1.73 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr11_+_101623776 | 1.73 |
ENSMUST00000039152.14
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr17_-_74630882 | 1.71 |
ENSMUST00000164832.9
|
Dpy30
|
dpy-30, histone methyltransferase complex regulatory subunit |
| chrX_+_20570145 | 1.69 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chrX_-_100463395 | 1.66 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chrX_+_93278588 | 1.61 |
ENSMUST00000096369.10
ENSMUST00000113911.9 |
Klhl15
|
kelch-like 15 |
| chrX_-_85348699 | 1.58 |
ENSMUST00000096378.3
|
Mageb4
|
MAGE family member B4 |
| chrX_-_85299825 | 1.57 |
ENSMUST00000113969.10
|
Mageb4
|
MAGE family member B4 |
| chr11_-_50718508 | 1.56 |
ENSMUST00000109135.9
|
Zfp354c
|
zinc finger protein 354C |
| chrX_-_139714182 | 1.54 |
ENSMUST00000044179.8
|
Tex13b
|
testis expressed 13B |
| chr7_-_140793990 | 1.53 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chrX_+_134643435 | 1.52 |
ENSMUST00000096321.3
ENSMUST00000113144.8 ENSMUST00000113147.8 ENSMUST00000113145.8 |
Armcx5
Gprasp1
|
armadillo repeat containing, X-linked 5 G protein-coupled receptor associated sorting protein 1 |
| chr7_-_126062272 | 1.52 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr1_-_156766351 | 1.51 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chrX_+_135567124 | 1.51 |
ENSMUST00000060904.11
ENSMUST00000113100.2 ENSMUST00000128040.2 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr2_-_32665596 | 1.49 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr17_-_35454729 | 1.47 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr7_+_6048160 | 1.46 |
ENSMUST00000037728.13
ENSMUST00000121583.2 |
Nlrp4c
|
NLR family, pyrin domain containing 4C |
| chr4_+_146586445 | 1.46 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr17_+_48653493 | 1.43 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr9_-_20553576 | 1.42 |
ENSMUST00000155301.8
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr2_+_156563284 | 1.39 |
ENSMUST00000099145.6
|
Dlgap4
|
DLG associated protein 4 |
| chr10_-_127016448 | 1.39 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr17_+_48653429 | 1.37 |
ENSMUST00000024791.15
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chrX_-_72442342 | 1.36 |
ENSMUST00000180787.3
|
Gm18336
|
predicted gene, 18336 |
| chr5_+_122988111 | 1.36 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chr6_-_83549399 | 1.35 |
ENSMUST00000206592.2
ENSMUST00000206400.2 |
Stambp
|
STAM binding protein |
| chr7_-_142211203 | 1.33 |
ENSMUST00000097936.9
ENSMUST00000000033.12 |
Igf2
|
insulin-like growth factor 2 |
| chr19_-_46314945 | 1.31 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr18_+_34758062 | 1.28 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr2_-_3475990 | 1.25 |
ENSMUST00000060618.13
|
Suv39h2
|
suppressor of variegation 3-9 2 |
| chr1_-_25868788 | 1.24 |
ENSMUST00000151309.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr7_-_126398165 | 1.22 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr1_+_40123858 | 1.21 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr1_-_156766381 | 1.20 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr5_+_110324734 | 1.18 |
ENSMUST00000139611.8
ENSMUST00000031477.9 |
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr7_-_126398343 | 1.16 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr18_+_34757687 | 1.16 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr15_-_3333003 | 1.13 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr17_+_37269513 | 1.11 |
ENSMUST00000173814.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr2_-_152672535 | 1.09 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr7_-_130964469 | 1.07 |
ENSMUST00000059438.11
|
2310057M21Rik
|
RIKEN cDNA 2310057M21 gene |
| chr11_-_113956996 | 1.06 |
ENSMUST00000041627.14
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr18_+_34757666 | 1.04 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr5_+_37892863 | 1.04 |
ENSMUST00000073554.4
|
Cytl1
|
cytokine-like 1 |
| chr16_+_4544221 | 1.04 |
ENSMUST00000154117.2
ENSMUST00000004172.15 |
Hmox2
|
heme oxygenase 2 |
| chr1_-_156766957 | 1.02 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr12_-_85017586 | 1.00 |
ENSMUST00000165886.2
ENSMUST00000167448.8 ENSMUST00000043169.14 |
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr7_-_35346889 | 1.00 |
ENSMUST00000118501.8
|
Pdcd5
|
programmed cell death 5 |
| chr11_+_101623836 | 0.96 |
ENSMUST00000129741.2
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr5_+_140404997 | 0.92 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr3_+_66127330 | 0.90 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr1_-_25868592 | 0.88 |
ENSMUST00000135518.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr7_-_4973960 | 0.87 |
ENSMUST00000144863.8
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr7_-_35346553 | 0.86 |
ENSMUST00000120714.2
|
Pdcd5
|
programmed cell death 5 |
| chr17_+_35278011 | 0.85 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr17_+_37269468 | 0.85 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr8_-_84903181 | 0.84 |
ENSMUST00000070102.6
ENSMUST00000210202.2 |
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr6_+_88701578 | 0.83 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr5_-_132570710 | 0.82 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr11_-_102047165 | 0.81 |
ENSMUST00000021296.7
|
Tmem101
|
transmembrane protein 101 |
| chr10_+_34173426 | 0.80 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr8_+_72021510 | 0.80 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr11_-_58346806 | 0.80 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr3_+_145926709 | 0.80 |
ENSMUST00000039164.4
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr9_-_123680726 | 0.79 |
ENSMUST00000084715.14
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr17_-_56343625 | 0.78 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr7_+_25016492 | 0.77 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr5_+_137286535 | 0.75 |
ENSMUST00000024099.11
ENSMUST00000196208.5 ENSMUST00000085934.4 |
Ache
|
acetylcholinesterase |
| chr3_-_36667610 | 0.74 |
ENSMUST00000108156.9
|
Bbs7
|
Bardet-Biedl syndrome 7 (human) |
| chr2_-_32665637 | 0.73 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr9_+_56979307 | 0.73 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr15_-_11907267 | 0.72 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chr5_+_110324506 | 0.72 |
ENSMUST00000112512.8
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr14_-_70879694 | 0.69 |
ENSMUST00000227123.2
ENSMUST00000022697.7 |
Fgf17
|
fibroblast growth factor 17 |
| chr5_+_27466914 | 0.66 |
ENSMUST00000101471.4
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr3_+_67799510 | 0.66 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr12_-_101924407 | 0.66 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr13_-_22689551 | 0.65 |
ENSMUST00000228020.2
|
Vmn1r202
|
vomeronasal 1 receptor 202 |
| chr15_-_101422054 | 0.65 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr11_-_32217547 | 0.64 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr7_-_142215595 | 0.62 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr2_+_30331839 | 0.49 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr2_-_131170902 | 0.48 |
ENSMUST00000110194.8
|
Rnf24
|
ring finger protein 24 |
| chr1_-_151304191 | 0.46 |
ENSMUST00000064771.12
|
Swt1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr15_+_92495007 | 0.46 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr7_+_138980405 | 0.46 |
ENSMUST00000097975.3
|
Inpp5a
|
inositol polyphosphate-5-phosphatase A |
| chr2_-_31031814 | 0.44 |
ENSMUST00000073879.12
ENSMUST00000100208.9 ENSMUST00000100207.9 ENSMUST00000113555.8 ENSMUST00000075326.11 ENSMUST00000113552.9 ENSMUST00000136181.8 ENSMUST00000113564.9 ENSMUST00000113562.9 ENSMUST00000113560.8 |
Fnbp1
|
formin binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 1.5 | 12.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.3 | 5.4 | GO:2000506 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 1.0 | 4.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.0 | 5.8 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.9 | 2.8 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.8 | 2.3 | GO:2000474 | cellular response to morphine(GO:0071315) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.8 | 5.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.7 | 3.0 | GO:0071895 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.7 | 2.0 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.6 | 3.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 4.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.5 | 1.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.5 | 4.7 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.5 | 2.7 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.4 | 1.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 3.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 2.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.4 | 3.6 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 5.9 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.4 | 8.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 3.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 4.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 1.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 1.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 2.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.3 | 1.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 4.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.3 | 1.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.3 | 0.9 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 8.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.3 | 3.7 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.2 | 0.7 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 1.0 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 3.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 4.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 0.6 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.2 | 1.9 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.2 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 2.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.8 | GO:0097155 | embryonic heart tube left/right pattern formation(GO:0060971) fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 2.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 3.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 3.0 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 0.5 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 2.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 7.9 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 0.6 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 2.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.9 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 1.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 2.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 2.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 1.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 2.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 8.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.4 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 1.5 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.7 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 1.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 2.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.1 | GO:0050883 | vestibular nucleus development(GO:0021750) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 5.1 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 6.5 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 35.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 3.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 2.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 1.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 2.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 4.9 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 7.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0070099 | regulation of chemokine-mediated signaling pathway(GO:0070099) |
| 0.0 | 0.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 3.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 2.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 1.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 3.9 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.6 | 8.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.6 | 4.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.5 | 4.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.4 | 7.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 2.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 3.5 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 8.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 1.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 2.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 6.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 4.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 4.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.2 | 3.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 3.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 2.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 3.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 3.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 19.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 2.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 5.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.1 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 4.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 4.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 7.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 8.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 2.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 2.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 8.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 10.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 1.1 | 8.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.0 | 4.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.8 | 5.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.6 | 1.9 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.6 | 3.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.6 | 3.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 3.7 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.5 | 5.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 3.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 2.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.4 | 2.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 1.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 2.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.3 | 1.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 0.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 5.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 3.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 3.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 4.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 2.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 1.3 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 7.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 4.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 4.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.5 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 2.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 8.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 4.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 7.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 3.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 7.6 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 4.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 4.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 4.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 2.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 3.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 13.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 4.3 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 3.9 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 3.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 5.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 2.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 3.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 5.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 6.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 5.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 4.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |