Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
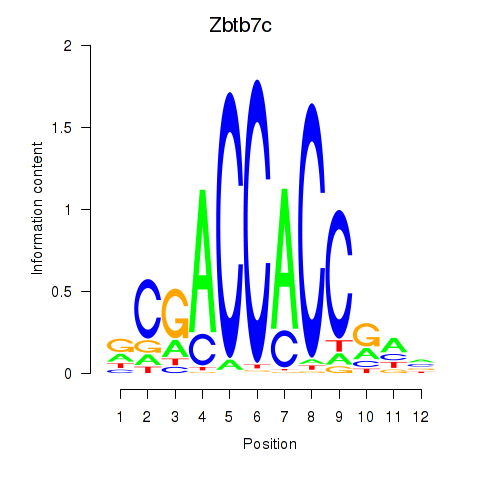
Results for Zbtb7c
Z-value: 1.37
Transcription factors associated with Zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7c
|
ENSMUSG00000044646.16 | Zbtb7c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7c | mm39_v1_chr18_+_75953244_75953308 | 0.11 | 3.6e-01 | Click! |
Activity profile of Zbtb7c motif
Sorted Z-values of Zbtb7c motif
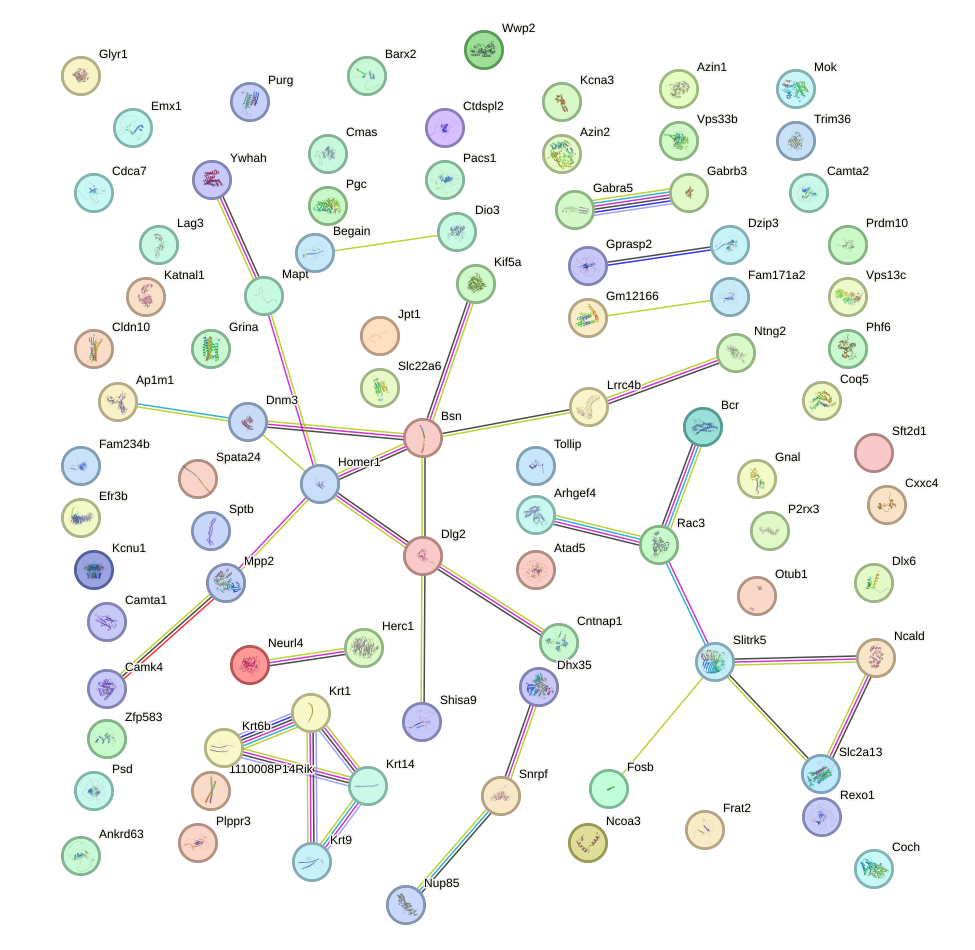
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_127099183 | 34.39 |
ENSMUST00000099172.5
|
Kif5a
|
kinesin family member 5A |
| chr10_-_127098932 | 32.65 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr18_+_33072194 | 16.23 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr7_-_57159119 | 11.23 |
ENSMUST00000206382.2
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr12_-_109034099 | 10.27 |
ENSMUST00000190647.3
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr11_-_100084072 | 9.96 |
ENSMUST00000059707.3
|
Krt9
|
keratin 9 |
| chr14_+_111912529 | 9.79 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr19_-_46306506 | 8.91 |
ENSMUST00000224556.2
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr12_-_4088905 | 8.39 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chr6_+_135174975 | 8.33 |
ENSMUST00000111915.8
ENSMUST00000111916.2 |
Fam234b
|
family with sequence similarity 234, member B |
| chrX_+_134739783 | 8.06 |
ENSMUST00000173804.8
ENSMUST00000113136.8 |
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr15_-_101759212 | 7.78 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr16_+_11802445 | 7.44 |
ENSMUST00000170672.9
ENSMUST00000023138.8 |
Shisa9
|
shisa family member 9 |
| chr7_+_44091822 | 7.43 |
ENSMUST00000058667.15
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr7_+_57240250 | 7.29 |
ENSMUST00000196198.5
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr2_+_157401998 | 6.71 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr16_-_48814437 | 6.67 |
ENSMUST00000121869.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr1_+_34840785 | 6.62 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr6_+_135175031 | 6.44 |
ENSMUST00000130612.2
|
Fam234b
|
family with sequence similarity 234, member B |
| chr11_+_53410697 | 6.28 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr16_-_48814294 | 6.23 |
ENSMUST00000114516.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr15_+_78783867 | 6.21 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr11_+_101066867 | 6.17 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr1_-_162305573 | 5.92 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr7_-_19043955 | 5.90 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chr6_-_124888643 | 5.83 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr14_+_119092107 | 5.75 |
ENSMUST00000100314.4
|
Cldn10
|
claudin 10 |
| chr3_+_106943472 | 5.51 |
ENSMUST00000052718.5
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr15_-_37734579 | 5.47 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
| chr9_-_108067552 | 5.29 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr11_+_53410552 | 5.13 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr6_+_85164420 | 4.83 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr7_+_5059703 | 4.81 |
ENSMUST00000208042.2
ENSMUST00000207974.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr7_+_57240894 | 4.62 |
ENSMUST00000039697.14
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr5_+_33176160 | 4.60 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr19_-_7183596 | 4.47 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr6_+_6863269 | 4.24 |
ENSMUST00000171311.8
ENSMUST00000160937.9 |
Dlx6
|
distal-less homeobox 6 |
| chr10_-_79710067 | 4.19 |
ENSMUST00000166023.2
ENSMUST00000167707.2 ENSMUST00000165601.8 |
Plppr3
|
phospholipid phosphatase related 3 |
| chr19_-_7183626 | 4.05 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr9_+_66257747 | 3.98 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr9_+_31191820 | 3.91 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr5_+_136937035 | 3.86 |
ENSMUST00000199101.5
ENSMUST00000200153.5 |
Ift22
|
intraflagellar transport 22 |
| chr7_+_5059855 | 3.79 |
ENSMUST00000208161.2
ENSMUST00000207215.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr10_-_79710044 | 3.75 |
ENSMUST00000167897.8
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr10_-_80382611 | 3.64 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr7_+_5060159 | 3.56 |
ENSMUST00000045543.8
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr8_+_4216556 | 3.39 |
ENSMUST00000239400.2
ENSMUST00000177053.8 ENSMUST00000176149.9 ENSMUST00000176072.9 ENSMUST00000176825.3 |
Evi5l
|
ecotropic viral integration site 5 like |
| chr7_+_90125890 | 3.37 |
ENSMUST00000208919.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr13_+_93441307 | 3.35 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr2_+_72306503 | 3.24 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr11_-_102338473 | 3.22 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr12_+_51640097 | 3.15 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr8_+_26401698 | 2.92 |
ENSMUST00000120653.8
ENSMUST00000126226.2 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr5_-_148865429 | 2.85 |
ENSMUST00000149169.3
ENSMUST00000047257.15 |
Katnal1
|
katanin p60 subunit A-like 1 |
| chr15_+_78784043 | 2.84 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr12_+_102094977 | 2.81 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr11_+_79980210 | 2.79 |
ENSMUST00000017694.7
ENSMUST00000108239.7 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr8_+_33876353 | 2.69 |
ENSMUST00000070340.6
ENSMUST00000078058.5 |
Purg
|
purine-rich element binding protein G |
| chr11_-_115405200 | 2.60 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr11_+_75422516 | 2.58 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr19_-_5323092 | 2.54 |
ENSMUST00000237463.2
ENSMUST00000025786.9 |
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr2_-_32271833 | 2.51 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr5_+_30868908 | 2.51 |
ENSMUST00000114729.8
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr15_+_76130947 | 2.46 |
ENSMUST00000229772.2
ENSMUST00000230347.2 ENSMUST00000023225.8 |
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr9_+_67747668 | 2.35 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chrX_+_52001108 | 2.24 |
ENSMUST00000078944.13
ENSMUST00000101587.10 ENSMUST00000154864.4 |
Phf6
|
PHD finger protein 6 |
| chr15_-_38518458 | 2.23 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr7_-_6334239 | 2.22 |
ENSMUST00000127658.2
ENSMUST00000062765.14 |
Zfp583
|
zinc finger protein 583 |
| chr5_+_30869193 | 2.21 |
ENSMUST00000088081.11
ENSMUST00000101442.4 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr11_-_70578775 | 2.16 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr7_+_135139542 | 2.15 |
ENSMUST00000073961.8
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr5_+_136937083 | 2.06 |
ENSMUST00000008131.10
|
Ift22
|
intraflagellar transport 22 |
| chr6_+_6863769 | 2.02 |
ENSMUST00000031768.8
|
Dlx6
|
distal-less homeobox 6 |
| chr19_+_41818409 | 2.02 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas |
| chr7_-_141456092 | 1.92 |
ENSMUST00000055819.13
ENSMUST00000001950.12 |
Tollip
|
toll interacting protein |
| chr11_+_120612278 | 1.91 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr12_-_76756772 | 1.89 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr4_-_151946219 | 1.83 |
ENSMUST00000097774.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr2_-_118534444 | 1.79 |
ENSMUST00000104937.2
|
Ankrd63
|
ankyrin repeat domain 63 |
| chr9_-_31824758 | 1.72 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr1_-_16727067 | 1.70 |
ENSMUST00000188641.7
|
Eloc
|
elongin C |
| chr7_-_141456045 | 1.67 |
ENSMUST00000130439.3
|
Tollip
|
toll interacting protein |
| chr11_+_75422925 | 1.65 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr17_+_48037758 | 1.65 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr18_+_67266784 | 1.64 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr6_+_142702403 | 1.61 |
ENSMUST00000032419.9
|
Cmas
|
cytidine monophospho-N-acetylneuraminic acid synthetase |
| chr2_+_158636727 | 1.61 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr16_+_48814548 | 1.59 |
ENSMUST00000117994.8
ENSMUST00000048374.6 |
Cip2a
|
cell proliferation regulating inhibitor of protein phosphatase 2A |
| chr11_+_120612369 | 1.58 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr2_-_29142965 | 1.58 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr12_+_110245662 | 1.53 |
ENSMUST00000097228.5
|
Dio3
|
deiodinase, iodothyronine type III |
| chr11_-_70578905 | 1.52 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr15_+_76131020 | 1.49 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr11_+_75422953 | 1.46 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr17_+_8529932 | 1.44 |
ENSMUST00000154553.2
ENSMUST00000140890.3 |
Sft2d1
Gm49987
|
SFT2 domain containing 1 predicted gene, 49987 |
| chr12_-_110807330 | 1.42 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr19_+_8595369 | 1.41 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_-_92231517 | 1.40 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr15_-_38518406 | 1.38 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr18_-_46345661 | 1.36 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr7_+_79939747 | 1.17 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr18_-_35795233 | 1.16 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr10_+_74896383 | 1.16 |
ENSMUST00000164107.3
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chr8_+_72993862 | 1.15 |
ENSMUST00000003117.15
ENSMUST00000212841.2 |
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr11_-_70578744 | 1.09 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr4_-_128856213 | 1.06 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr11_+_104122399 | 1.05 |
ENSMUST00000132977.8
ENSMUST00000132245.8 ENSMUST00000100347.11 |
Mapt
|
microtubule-associated protein tau |
| chr16_-_4867703 | 0.99 |
ENSMUST00000115844.3
ENSMUST00000023189.15 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr2_+_121786892 | 0.96 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr10_-_93425553 | 0.92 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr3_+_133942244 | 0.83 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr19_-_41836514 | 0.80 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr15_-_101588714 | 0.79 |
ENSMUST00000023786.7
|
Krt6b
|
keratin 6B |
| chr8_+_108162985 | 0.78 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_+_69792642 | 0.78 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr11_+_104122341 | 0.77 |
ENSMUST00000106993.10
|
Mapt
|
microtubule-associated protein tau |
| chr2_+_121786444 | 0.75 |
ENSMUST00000036647.13
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr2_-_84865831 | 0.74 |
ENSMUST00000028465.14
|
P2rx3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr18_-_35795175 | 0.73 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr5_+_115417725 | 0.71 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr11_+_115455260 | 0.67 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr11_-_100098333 | 0.66 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr11_+_69656725 | 0.65 |
ENSMUST00000108640.8
ENSMUST00000108639.8 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr4_-_126150066 | 0.59 |
ENSMUST00000122129.8
ENSMUST00000061143.15 ENSMUST00000106132.3 |
Map7d1
|
MAP7 domain containing 1 |
| chr1_-_16727242 | 0.49 |
ENSMUST00000186948.7
ENSMUST00000187910.7 ENSMUST00000115352.10 |
Eloc
|
elongin C |
| chr1_-_16727133 | 0.44 |
ENSMUST00000185771.7
|
Eloc
|
elongin C |
| chr10_+_84938452 | 0.40 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr1_-_106641940 | 0.40 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr11_+_68322945 | 0.39 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr11_+_69656797 | 0.36 |
ENSMUST00000108642.8
ENSMUST00000156932.8 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr5_-_134643805 | 0.35 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr11_+_70453806 | 0.34 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr1_-_171188302 | 0.23 |
ENSMUST00000061878.5
|
Klhdc9
|
kelch domain containing 9 |
| chr2_+_174602574 | 0.23 |
ENSMUST00000140908.2
|
Edn3
|
endothelin 3 |
| chr7_+_79992839 | 0.20 |
ENSMUST00000032747.7
ENSMUST00000206480.2 ENSMUST00000206074.2 ENSMUST00000206122.2 |
Hddc3
|
HD domain containing 3 |
| chr16_-_22084700 | 0.19 |
ENSMUST00000161286.8
|
Tra2b
|
transformer 2 beta |
| chr7_+_35285657 | 0.19 |
ENSMUST00000040844.16
ENSMUST00000188906.7 ENSMUST00000186245.7 ENSMUST00000190503.7 |
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr5_+_108416763 | 0.16 |
ENSMUST00000031190.5
|
Dr1
|
down-regulator of transcription 1 |
| chr2_+_119181703 | 0.16 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr8_+_72993913 | 0.14 |
ENSMUST00000145213.8
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr2_-_121786573 | 0.12 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr15_-_101602734 | 0.05 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr11_-_70895213 | 0.02 |
ENSMUST00000124464.2
ENSMUST00000108527.8 |
Dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr13_-_30168374 | 0.01 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.4 | 67.0 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 2.0 | 5.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 1.8 | 5.3 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.5 | 4.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.2 | 4.7 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.8 | 2.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 7.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.6 | 16.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.6 | 6.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 2.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.5 | 11.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 4.6 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.4 | 2.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.4 | 1.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 9.8 | GO:0021756 | striatum development(GO:0021756) |
| 0.3 | 3.5 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.3 | 1.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 7.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 4.8 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 8.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 8.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 4.0 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 8.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 8.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 1.6 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 3.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 2.8 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 | 3.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 12.1 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.2 | 1.5 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.2 | 1.5 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.1 | 0.7 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 3.9 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 1.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 3.6 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 7.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.7 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 0.4 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) positive regulation of neuron maturation(GO:0014042) nitrogen utilization(GO:0019740) |
| 0.1 | 2.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 5.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 7.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.4 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 2.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 3.0 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 5.7 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 1.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 5.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 7.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 6.7 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 5.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 3.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 9.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 6.0 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 1.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 9.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 3.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 67.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 2.0 | 5.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.8 | 5.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.6 | 12.9 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.8 | 23.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.6 | 7.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 2.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.4 | 18.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 7.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 3.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 5.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 6.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 0.9 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 8.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 16.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 8.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 4.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 3.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 10.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 14.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 67.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 2.0 | 5.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.3 | 7.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.2 | 8.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.2 | 11.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.2 | 4.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 1.0 | 16.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.0 | 5.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 3.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.7 | 12.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 7.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 11.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 2.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 2.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 1.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.4 | 1.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 2.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.0 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.3 | 1.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 3.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 5.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 8.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 5.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 6.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 3.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 5.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 8.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 3.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 5.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 15.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 8.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 7.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 3.6 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 3.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 6.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 5.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 4.7 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 3.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.8 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 5.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 6.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 6.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 6.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 67.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.0 | 23.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 3.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 5.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 16.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 3.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 4.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 5.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 6.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 13.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 6.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |