Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zfhx3
Z-value: 0.97
Transcription factors associated with Zfhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfhx3
|
ENSMUSG00000038872.11 | Zfhx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | mm39_v1_chr8_+_108669276_108669276 | -0.13 | 2.9e-01 | Click! |
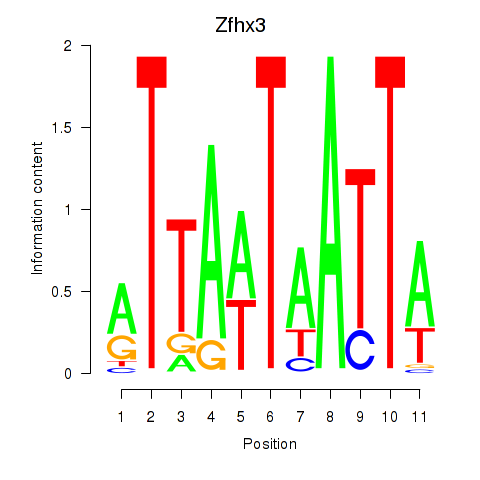
Activity profile of Zfhx3 motif
Sorted Z-values of Zfhx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfhx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_98919183 | 29.70 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr19_+_30210320 | 13.14 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr3_+_82933383 | 12.80 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr2_-_34951443 | 12.68 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr15_-_96929086 | 12.23 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr17_+_12597490 | 12.11 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr2_-_34990689 | 11.66 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr13_-_56696222 | 10.03 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_+_172525613 | 9.91 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr15_-_60793115 | 9.87 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr6_-_141892517 | 9.78 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr1_-_172722589 | 9.13 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr7_+_30193047 | 8.54 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr9_-_70841881 | 8.52 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr9_-_70842090 | 7.87 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr4_-_6275629 | 7.66 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr11_+_101258368 | 7.57 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr1_+_88093726 | 7.49 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr13_-_56696310 | 7.15 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_137923521 | 6.54 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_+_24023428 | 5.65 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr11_+_70410445 | 4.98 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr13_-_24098981 | 4.68 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr11_+_70410009 | 4.35 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr2_-_25517945 | 4.34 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr13_-_24098951 | 4.33 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr5_+_35198853 | 4.15 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr2_-_134396268 | 3.91 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr3_-_137837117 | 3.88 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr13_+_24023386 | 3.78 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chrM_+_9870 | 3.70 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr17_-_79292856 | 3.57 |
ENSMUST00000118991.2
|
Prkd3
|
protein kinase D3 |
| chr7_+_51537645 | 3.53 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr2_+_162829250 | 3.38 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_-_38649291 | 3.38 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr17_+_79919267 | 3.10 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_+_162829422 | 2.90 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr11_+_78356523 | 2.76 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr19_-_46661321 | 2.60 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr4_+_150938376 | 2.48 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr15_+_99291455 | 2.37 |
ENSMUST00000162624.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr12_+_59142439 | 2.34 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr15_+_99291491 | 2.33 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_+_31224616 | 2.27 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr19_-_46661501 | 2.23 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr15_+_31225302 | 2.20 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr18_-_66155651 | 2.14 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr15_+_31224460 | 2.13 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chrM_+_9459 | 1.99 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr18_+_36414122 | 1.95 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr4_+_138694422 | 1.87 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chr11_+_114566257 | 1.84 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr15_+_31224555 | 1.73 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr2_+_152873772 | 1.69 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr5_-_28672091 | 1.67 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr6_+_113460258 | 1.63 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr14_+_26722319 | 1.56 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr7_-_99629637 | 1.52 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr2_+_120807498 | 1.36 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr3_+_85946145 | 1.18 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chrM_+_14138 | 1.16 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr1_-_80439165 | 1.14 |
ENSMUST00000211023.2
|
Gm45261
|
predicted gene 45261 |
| chr2_-_63014514 | 1.11 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chrX_-_142716085 | 1.09 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr2_+_85868891 | 1.09 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr2_+_152907875 | 1.05 |
ENSMUST00000238488.2
ENSMUST00000129377.8 ENSMUST00000109800.2 |
Ccm2l
|
cerebral cavernous malformation 2-like |
| chr17_-_37611375 | 1.04 |
ENSMUST00000058046.6
|
Olfr101
|
olfactory receptor 101 |
| chr2_-_140513382 | 1.02 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chrM_+_7006 | 1.02 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr2_+_81883566 | 1.00 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr4_-_42168603 | 1.00 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_-_42665763 | 1.00 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr6_-_93769426 | 0.90 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_-_126758520 | 0.86 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr3_-_98364359 | 0.84 |
ENSMUST00000188356.3
ENSMUST00000167753.8 |
Gm4450
|
predicted gene 4450 |
| chr14_+_75368939 | 0.83 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr18_+_36661198 | 0.81 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr8_-_68270936 | 0.80 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_-_12829100 | 0.79 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr7_-_101486983 | 0.79 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr15_+_39255185 | 0.78 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_63014622 | 0.76 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr14_-_30973164 | 0.75 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chrX_-_105647282 | 0.74 |
ENSMUST00000113480.2
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr2_-_140513320 | 0.74 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr1_-_126758369 | 0.73 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr17_+_38143840 | 0.71 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr13_-_18556626 | 0.70 |
ENSMUST00000139064.10
ENSMUST00000175703.9 |
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr11_-_99482165 | 0.69 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr9_+_7571397 | 0.67 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chrX_+_111150171 | 0.66 |
ENSMUST00000164272.3
ENSMUST00000132037.2 |
4933403O08Rik
|
RIKEN cDNA 4933403O08 gene |
| chr14_+_33073249 | 0.64 |
ENSMUST00000140711.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr7_+_18853778 | 0.59 |
ENSMUST00000053109.5
|
Fbxo46
|
F-box protein 46 |
| chr2_-_165210622 | 0.58 |
ENSMUST00000141140.2
ENSMUST00000103085.8 |
Zfp663
|
zinc finger protein 663 |
| chr2_+_69727599 | 0.58 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_69727563 | 0.57 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr16_+_16688692 | 0.57 |
ENSMUST00000232547.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chrX_+_159551171 | 0.55 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr16_-_56748424 | 0.53 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr14_+_75368532 | 0.53 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_-_86218905 | 0.51 |
ENSMUST00000213998.2
|
Olfr1058
|
olfactory receptor 1058 |
| chr5_+_117378510 | 0.51 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr2_+_87725306 | 0.50 |
ENSMUST00000217436.2
|
Olfr1153
|
olfactory receptor 1153 |
| chr4_+_109092829 | 0.48 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr7_-_107633196 | 0.48 |
ENSMUST00000210173.3
|
Olfr478
|
olfactory receptor 478 |
| chrX_+_159551009 | 0.46 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr7_+_102420428 | 0.46 |
ENSMUST00000213432.2
|
Olfr561
|
olfactory receptor 561 |
| chr10_-_129593612 | 0.46 |
ENSMUST00000213379.2
ENSMUST00000217106.2 |
Olfr807
|
olfactory receptor 807 |
| chrX_-_110446022 | 0.44 |
ENSMUST00000156639.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr10_-_56104732 | 0.43 |
ENSMUST00000099739.5
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr1_+_179788037 | 0.42 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_+_48906825 | 0.42 |
ENSMUST00000031837.8
|
Doxl1
|
diamine oxidase-like protein 1 |
| chr4_+_109092610 | 0.39 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr10_-_129591354 | 0.37 |
ENSMUST00000059038.3
|
Olfr807
|
olfactory receptor 807 |
| chr14_-_50476340 | 0.36 |
ENSMUST00000059565.2
|
Olfr731
|
olfactory receptor 731 |
| chr7_-_10011933 | 0.36 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr7_+_102834996 | 0.36 |
ENSMUST00000218618.2
|
Olfr592
|
olfactory receptor 592 |
| chr6_+_21986445 | 0.31 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_66672158 | 0.28 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_-_70560110 | 0.28 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr7_-_139953579 | 0.27 |
ENSMUST00000074177.3
|
Olfr530
|
olfactory receptor 530 |
| chr4_+_109092459 | 0.25 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr5_-_87638728 | 0.23 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr8_+_34006758 | 0.23 |
ENSMUST00000149399.8
|
Tex15
|
testis expressed gene 15 |
| chr5_-_86521273 | 0.23 |
ENSMUST00000031175.12
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr3_+_108479015 | 0.22 |
ENSMUST00000143054.2
|
Taf13
|
TATA-box binding protein associated factor 13 |
| chr10_-_107747995 | 0.21 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr1_-_4430481 | 0.20 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_-_72817060 | 0.20 |
ENSMUST00000112062.2
|
Gm11084
|
predicted gene 11084 |
| chrX_-_142716200 | 0.19 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr1_-_157084252 | 0.17 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr7_+_105017437 | 0.17 |
ENSMUST00000098152.3
ENSMUST00000217827.2 |
Olfr692
|
olfactory receptor 692 |
| chr2_-_111880531 | 0.17 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr4_+_115741581 | 0.16 |
ENSMUST00000097918.3
|
Kncn
|
kinocilin |
| chrX_+_71016592 | 0.15 |
ENSMUST00000066116.8
|
Fate1
|
fetal and adult testis expressed 1 |
| chr6_+_132886614 | 0.15 |
ENSMUST00000076119.6
|
Tas2r125
|
taste receptor, type 2, member 125 |
| chrM_-_14061 | 0.14 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr18_+_56695515 | 0.14 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr7_+_104274001 | 0.13 |
ENSMUST00000215454.3
ENSMUST00000213297.2 |
Olfr657
|
olfactory receptor 657 |
| chr9_-_48876290 | 0.12 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr3_-_103553347 | 0.12 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr7_-_28661751 | 0.12 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr3_+_125197722 | 0.09 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr19_+_48194464 | 0.09 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr5_-_137784912 | 0.07 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr7_+_4340708 | 0.05 |
ENSMUST00000006792.6
ENSMUST00000126417.3 |
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr2_-_89195205 | 0.03 |
ENSMUST00000111543.2
ENSMUST00000137692.3 |
Olfr1234
|
olfactory receptor 1234 |
| chr19_+_58500411 | 0.02 |
ENSMUST00000235305.2
ENSMUST00000069419.8 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr10_+_90412114 | 0.01 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 29.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 3.3 | 16.4 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 3.2 | 12.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 3.0 | 9.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 2.8 | 8.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.5 | 12.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.4 | 12.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.6 | 6.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.5 | 7.7 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 1.3 | 17.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.2 | 9.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.9 | 4.7 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.9 | 20.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.8 | 3.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 7.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.6 | 1.7 | GO:1904339 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.5 | 3.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 3.9 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 2.5 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.4 | 2.8 | GO:0015886 | heme transport(GO:0015886) methotrexate transport(GO:0051958) |
| 0.3 | 9.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 1.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 12.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 4.8 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 1.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 1.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 1.2 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 8.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.5 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 4.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 9.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 2.3 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 11.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 5.7 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 2.0 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 3.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.6 | 12.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.4 | 12.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 16.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 15.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 5.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 1.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 8.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 28.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 19.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 7.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 10.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 10.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 6.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.4 | GO:0035478 | chylomicron binding(GO:0035478) |
| 2.7 | 29.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 2.4 | 19.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.2 | 6.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.9 | 7.6 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.6 | 13.1 | GO:0005534 | galactose binding(GO:0005534) |
| 1.2 | 18.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.0 | 3.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.8 | 4.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.8 | 12.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 1.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 2.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 8.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 6.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 8.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 4.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 3.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 7.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 5.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 7.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 6.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 3.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.2 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 9.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 1.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 7.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 7.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 24.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 29.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 9.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 9.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 17.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 12.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 23.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.8 | 12.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 7.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 20.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 12.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.6 | 6.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 8.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 8.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 12.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 7.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 2.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |