Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
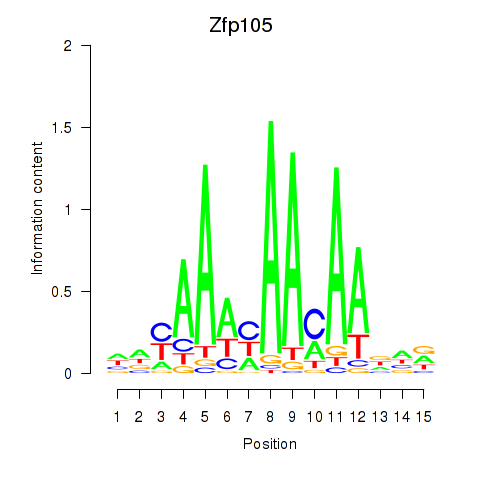
Results for Zfp105
Z-value: 0.73
Transcription factors associated with Zfp105
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp105
|
ENSMUSG00000057895.12 | Zfp105 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp105 | mm39_v1_chr9_+_122752116_122752157 | 0.31 | 8.3e-03 | Click! |
Activity profile of Zfp105 motif
Sorted Z-values of Zfp105 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp105
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_75093177 | 5.29 |
ENSMUST00000129281.8
ENSMUST00000148144.8 ENSMUST00000130384.2 |
Myo5a
|
myosin VA |
| chr10_-_30647836 | 4.76 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr6_-_142910094 | 4.50 |
ENSMUST00000032421.4
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr15_-_50753061 | 3.87 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr5_+_86219593 | 3.64 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr10_-_30647881 | 3.37 |
ENSMUST00000215740.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr6_+_135339543 | 3.21 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr6_+_135339929 | 2.77 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr3_+_63203235 | 2.56 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr6_+_29859372 | 2.47 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_-_53797748 | 2.40 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr2_+_103799873 | 2.36 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr15_+_6673167 | 2.34 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr11_+_44508137 | 2.32 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr10_-_111833138 | 2.16 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr4_+_74160705 | 2.11 |
ENSMUST00000077851.10
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr4_+_101407608 | 2.04 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr3_+_31150982 | 1.88 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr3_+_63203516 | 1.88 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr5_+_147456497 | 1.74 |
ENSMUST00000175807.8
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr15_-_50752437 | 1.74 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr8_+_94537910 | 1.65 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_-_66915756 | 1.62 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr10_-_78427721 | 1.50 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr8_-_57940834 | 1.32 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr2_+_74534959 | 1.23 |
ENSMUST00000151380.2
|
Hoxd8
|
homeobox D8 |
| chr5_+_17779273 | 1.22 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr6_-_127128007 | 1.20 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr12_+_3941728 | 1.20 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr6_-_127127959 | 1.19 |
ENSMUST00000201637.2
|
Ccnd2
|
cyclin D2 |
| chr7_-_15781838 | 1.19 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr6_-_127127993 | 1.12 |
ENSMUST00000201066.2
|
Ccnd2
|
cyclin D2 |
| chr7_-_79974166 | 1.09 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr18_+_69477541 | 1.03 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr5_+_146769700 | 0.98 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr6_-_99412306 | 0.97 |
ENSMUST00000113322.9
ENSMUST00000176850.8 ENSMUST00000176632.8 |
Foxp1
|
forkhead box P1 |
| chr6_+_97968737 | 0.97 |
ENSMUST00000043628.13
ENSMUST00000203938.2 |
Mitf
|
melanogenesis associated transcription factor |
| chr15_-_50753437 | 0.96 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_-_30390996 | 0.88 |
ENSMUST00000152391.9
ENSMUST00000115184.2 ENSMUST00000080812.14 ENSMUST00000102992.10 |
Zc3hc1
|
zinc finger, C3HC type 1 |
| chr7_-_132724889 | 0.85 |
ENSMUST00000166439.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr9_+_57444801 | 0.84 |
ENSMUST00000093833.6
ENSMUST00000114200.10 |
Fam219b
|
family with sequence similarity 219, member B |
| chr12_-_52018072 | 0.82 |
ENSMUST00000040583.7
|
Heatr5a
|
HEAT repeat containing 5A |
| chr9_+_7692087 | 0.78 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
| chr9_+_108437485 | 0.73 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr5_+_76288524 | 0.72 |
ENSMUST00000152642.8
ENSMUST00000127278.8 |
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chrX_+_7656225 | 0.66 |
ENSMUST00000136930.8
ENSMUST00000115675.9 ENSMUST00000101694.10 |
Gripap1
|
GRIP1 associated protein 1 |
| chr9_+_21867043 | 0.66 |
ENSMUST00000053583.7
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr10_-_23977810 | 0.64 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr18_+_69633741 | 0.62 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr11_+_83742961 | 0.58 |
ENSMUST00000146786.8
|
Hnf1b
|
HNF1 homeobox B |
| chr5_-_143133260 | 0.54 |
ENSMUST00000215102.2
ENSMUST00000213631.2 ENSMUST00000164536.5 |
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr6_+_21949986 | 0.47 |
ENSMUST00000149728.7
|
Ing3
|
inhibitor of growth family, member 3 |
| chr3_+_20043315 | 0.46 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chr2_-_151586063 | 0.45 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr9_-_117080869 | 0.44 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr12_+_87921198 | 0.33 |
ENSMUST00000110145.12
ENSMUST00000181843.2 ENSMUST00000180706.8 ENSMUST00000181394.8 ENSMUST00000181326.8 ENSMUST00000181300.2 |
Gm2042
|
predicted gene 2042 |
| chr2_-_65397850 | 0.33 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr5_+_129875821 | 0.27 |
ENSMUST00000171300.8
ENSMUST00000201874.4 |
Sumf2
|
sulfatase modifying factor 2 |
| chr7_-_34012934 | 0.26 |
ENSMUST00000206399.2
|
Garre1
|
granule associated Rac and RHOG effector 1 |
| chr11_+_101623836 | 0.26 |
ENSMUST00000129741.2
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr12_+_103564479 | 0.25 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr11_+_108811168 | 0.22 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr2_+_111329683 | 0.21 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr5_+_20112704 | 0.14 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr13_+_67052978 | 0.13 |
ENSMUST00000168767.9
|
Gm10767
|
predicted gene 10767 |
| chr2_-_87985537 | 0.11 |
ENSMUST00000216951.2
|
Olfr1167
|
olfactory receptor 1167 |
| chr12_+_87541491 | 0.06 |
ENSMUST00000220499.2
|
Eif1ad12
|
eukaryotic translation initiation factor 1A domain containing 12 |
| chr14_-_101846551 | 0.05 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr11_-_99742434 | 0.05 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr5_+_20112500 | 0.02 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_-_55103598 | 0.01 |
ENSMUST00000036412.4
|
Zpld1
|
zona pellucida like domain containing 1 |
| chr3_-_116388334 | 0.00 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 1.1 | 5.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.6 | 4.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 8.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 2.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 1.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 1.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.3 | 2.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 0.8 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.3 | 8.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 3.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 6.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 0.7 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.2 | 1.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 2.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 1.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.6 | GO:0061296 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 2.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.7 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.1 | 1.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 4.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 2.4 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 1.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 2.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.9 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.4 | 3.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 1.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 2.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 11.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.7 | 4.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 2.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 4.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 1.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.7 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 2.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 8.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 5.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 8.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 7.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 3.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 3.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 3.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |