Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zfp148
Z-value: 0.75
Transcription factors associated with Zfp148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp148
|
ENSMUSG00000022811.18 | Zfp148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp148 | mm39_v1_chr16_+_33201164_33201206 | 0.20 | 8.4e-02 | Click! |
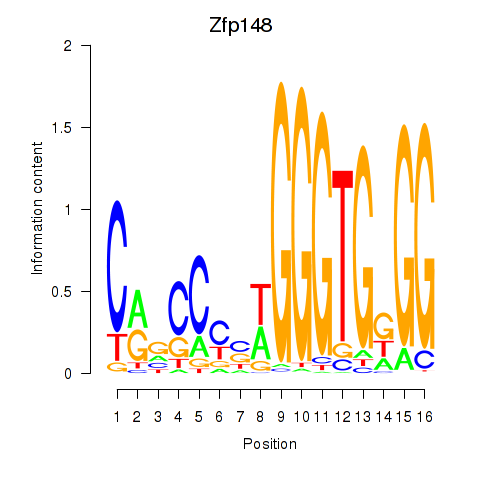
Activity profile of Zfp148 motif
Sorted Z-values of Zfp148 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp148
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_113115632 | 4.82 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr3_+_90520408 | 4.27 |
ENSMUST00000198128.2
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr12_+_113112311 | 4.19 |
ENSMUST00000199089.5
|
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr3_+_90520176 | 3.92 |
ENSMUST00000001051.9
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr11_+_72851989 | 3.53 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr1_+_135764092 | 3.25 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr11_+_117740077 | 3.05 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chrX_-_73290140 | 2.85 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr9_+_110848339 | 2.84 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr9_-_109678685 | 2.81 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr5_+_30745447 | 2.68 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr17_+_35268942 | 2.64 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr6_-_41681273 | 2.63 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr14_+_33807935 | 2.54 |
ENSMUST00000022519.15
|
Anxa8
|
annexin A8 |
| chr19_-_21449916 | 2.49 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr6_-_72935171 | 2.45 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr4_+_129948803 | 2.41 |
ENSMUST00000123617.8
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chrX_-_73289970 | 2.33 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr11_+_117740111 | 2.27 |
ENSMUST00000093906.5
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr4_+_114914880 | 2.26 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr6_+_29433247 | 2.22 |
ENSMUST00000101617.9
ENSMUST00000065090.8 |
Flnc
|
filamin C, gamma |
| chr3_+_90173813 | 2.15 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr5_+_8710059 | 2.14 |
ENSMUST00000047753.5
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr8_+_3715747 | 2.13 |
ENSMUST00000014118.4
|
Mcemp1
|
mast cell expressed membrane protein 1 |
| chr14_+_33807964 | 2.10 |
ENSMUST00000120077.2
|
Anxa8
|
annexin A8 |
| chr14_+_56003406 | 2.03 |
ENSMUST00000057569.4
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chrX_+_72760183 | 2.02 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr12_-_113271532 | 2.00 |
ENSMUST00000192188.3
ENSMUST00000103418.3 |
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr6_-_72935382 | 1.99 |
ENSMUST00000144337.2
|
Tmsb10
|
thymosin, beta 10 |
| chr7_-_80453033 | 1.95 |
ENSMUST00000167377.3
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr6_-_72935468 | 1.91 |
ENSMUST00000114050.8
|
Tmsb10
|
thymosin, beta 10 |
| chr17_-_32385826 | 1.89 |
ENSMUST00000087723.5
|
Notch3
|
notch 3 |
| chr5_+_122239007 | 1.89 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr5_+_122239030 | 1.88 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr9_-_107512511 | 1.85 |
ENSMUST00000192615.6
ENSMUST00000192837.2 ENSMUST00000193876.2 |
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr2_+_24275321 | 1.82 |
ENSMUST00000056641.15
ENSMUST00000142522.8 ENSMUST00000131930.2 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr18_+_4920513 | 1.78 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr10_-_81335966 | 1.70 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr9_-_107512566 | 1.68 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr5_+_122239830 | 1.65 |
ENSMUST00000146733.5
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr2_-_157046386 | 1.63 |
ENSMUST00000029170.8
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr2_+_103800459 | 1.59 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr10_+_79833296 | 1.59 |
ENSMUST00000171637.8
ENSMUST00000043866.8 |
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr2_+_103800553 | 1.58 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr17_-_37178079 | 1.58 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr7_+_28834276 | 1.57 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chrX_-_47543029 | 1.54 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr13_-_34186723 | 1.53 |
ENSMUST00000167237.8
ENSMUST00000168400.8 ENSMUST00000166354.2 ENSMUST00000076532.14 ENSMUST00000171034.8 |
Serpinb6a
|
serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| chr7_+_79944198 | 1.51 |
ENSMUST00000163812.9
ENSMUST00000047558.14 ENSMUST00000174199.8 ENSMUST00000173824.8 ENSMUST00000174172.8 |
Prc1
|
protein regulator of cytokinesis 1 |
| chr2_-_66615247 | 1.46 |
ENSMUST00000042792.7
|
Scn7a
|
sodium channel, voltage-gated, type VII, alpha |
| chr14_-_67953035 | 1.43 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr17_+_24072493 | 1.40 |
ENSMUST00000061725.8
|
Prss32
|
protease, serine 32 |
| chr17_-_24292453 | 1.40 |
ENSMUST00000017090.6
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr7_+_141276575 | 1.40 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr4_-_133480922 | 1.39 |
ENSMUST00000145664.9
ENSMUST00000105897.10 |
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr2_+_84670543 | 1.34 |
ENSMUST00000111624.8
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr11_-_84719779 | 1.34 |
ENSMUST00000047560.8
|
Dhrs11
|
dehydrogenase/reductase (SDR family) member 11 |
| chrX_+_133208833 | 1.31 |
ENSMUST00000081064.12
ENSMUST00000101251.8 ENSMUST00000129782.2 |
Cenpi
|
centromere protein I |
| chr3_-_90297187 | 1.31 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr3_+_95836558 | 1.29 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr13_+_73615316 | 1.28 |
ENSMUST00000022099.15
|
Lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr19_+_60878802 | 1.28 |
ENSMUST00000236876.2
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr2_+_84670956 | 1.27 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr10_+_3316057 | 1.26 |
ENSMUST00000043374.7
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr10_-_127031578 | 1.26 |
ENSMUST00000038217.14
ENSMUST00000130855.8 ENSMUST00000116229.2 ENSMUST00000144322.8 |
Dtx3
|
deltex 3, E3 ubiquitin ligase |
| chr3_+_95836637 | 1.25 |
ENSMUST00000171368.8
ENSMUST00000168106.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_+_89122499 | 1.25 |
ENSMUST00000142051.8
ENSMUST00000119084.2 |
Thbs3
|
thrombospondin 3 |
| chr7_+_138968988 | 1.24 |
ENSMUST00000106098.8
ENSMUST00000026550.14 |
Inpp5a
|
inositol polyphosphate-5-phosphatase A |
| chrX_+_72760318 | 1.24 |
ENSMUST00000114461.3
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr10_-_81360059 | 1.22 |
ENSMUST00000043709.8
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr4_+_134847949 | 1.22 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr5_-_137608886 | 1.22 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr15_-_99670479 | 1.21 |
ENSMUST00000023762.13
|
Cers5
|
ceramide synthase 5 |
| chr14_-_30348153 | 1.21 |
ENSMUST00000112211.9
ENSMUST00000112210.11 |
Prkcd
|
protein kinase C, delta |
| chr3_+_106393348 | 1.08 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr2_+_32496990 | 1.08 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr10_+_3316505 | 1.07 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chr7_-_132415257 | 1.07 |
ENSMUST00000097999.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr9_+_56344700 | 1.06 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr5_-_137608726 | 1.06 |
ENSMUST00000197912.5
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr11_-_53959758 | 1.05 |
ENSMUST00000093109.11
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr19_-_6835538 | 1.05 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr5_-_115439016 | 1.04 |
ENSMUST00000009157.4
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr15_-_99670276 | 1.04 |
ENSMUST00000109035.11
|
Cers5
|
ceramide synthase 5 |
| chr5_+_33815910 | 1.04 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_+_92524495 | 1.03 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr11_+_75542328 | 1.01 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr5_-_115438971 | 1.00 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr11_+_78234300 | 0.99 |
ENSMUST00000002127.14
ENSMUST00000108295.8 |
Unc119
|
unc-119 lipid binding chaperone |
| chr11_-_53959790 | 0.99 |
ENSMUST00000018755.10
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr11_+_4186391 | 0.98 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr4_-_127247864 | 0.97 |
ENSMUST00000106090.8
ENSMUST00000060419.2 |
Gjb4
|
gap junction protein, beta 4 |
| chr13_-_74956030 | 0.97 |
ENSMUST00000065629.6
|
Cast
|
calpastatin |
| chr11_+_75546671 | 0.97 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr4_+_148888877 | 0.97 |
ENSMUST00000094464.10
ENSMUST00000122222.8 |
Casz1
|
castor zinc finger 1 |
| chr2_+_84669739 | 0.96 |
ENSMUST00000146816.8
ENSMUST00000028469.14 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr11_+_69737200 | 0.95 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chr11_-_20781009 | 0.94 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr5_-_107873883 | 0.94 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr5_-_123270702 | 0.94 |
ENSMUST00000031401.6
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr7_-_25488060 | 0.92 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr11_+_96355413 | 0.92 |
ENSMUST00000103154.11
ENSMUST00000100521.10 ENSMUST00000100519.11 |
Skap1
|
src family associated phosphoprotein 1 |
| chr7_+_92524460 | 0.90 |
ENSMUST00000076052.8
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_-_3956991 | 0.90 |
ENSMUST00000126070.9
ENSMUST00000001801.11 |
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr15_-_99670751 | 0.88 |
ENSMUST00000175876.8
|
Cers5
|
ceramide synthase 5 |
| chr4_+_125918333 | 0.87 |
ENSMUST00000106162.8
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr15_-_103163860 | 0.86 |
ENSMUST00000075192.13
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr1_+_118317153 | 0.85 |
ENSMUST00000189738.7
ENSMUST00000187713.7 ENSMUST00000165223.8 ENSMUST00000189570.7 ENSMUST00000191445.7 ENSMUST00000189262.7 |
Clasp1
|
CLIP associating protein 1 |
| chr1_+_175708341 | 0.85 |
ENSMUST00000195196.6
ENSMUST00000194306.6 ENSMUST00000193822.6 |
Exo1
|
exonuclease 1 |
| chr4_+_59035088 | 0.85 |
ENSMUST00000041160.13
|
Gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr5_-_123270449 | 0.84 |
ENSMUST00000186469.7
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr7_+_28834350 | 0.83 |
ENSMUST00000159975.8
ENSMUST00000094617.11 ENSMUST00000032811.12 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr11_-_100830366 | 0.83 |
ENSMUST00000127638.8
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr19_+_6952319 | 0.82 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr12_+_71183622 | 0.81 |
ENSMUST00000149564.8
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr3_+_104696342 | 0.81 |
ENSMUST00000106787.8
ENSMUST00000176347.6 |
Rhoc
|
ras homolog family member C |
| chr1_-_131127825 | 0.81 |
ENSMUST00000068564.15
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr16_+_33504829 | 0.80 |
ENSMUST00000152782.8
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr3_+_104696108 | 0.79 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chr16_-_21982049 | 0.79 |
ENSMUST00000100052.11
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr11_-_101117237 | 0.78 |
ENSMUST00000100417.3
ENSMUST00000107285.8 ENSMUST00000107284.8 |
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr3_+_96604415 | 0.77 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr4_-_133695264 | 0.76 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_117688486 | 0.75 |
ENSMUST00000106331.2
|
6030468B19Rik
|
RIKEN cDNA 6030468B19 gene |
| chr7_+_96730915 | 0.75 |
ENSMUST00000206791.2
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr15_-_79287747 | 0.75 |
ENSMUST00000074991.10
|
Tmem184b
|
transmembrane protein 184b |
| chr10_-_76562002 | 0.75 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chr8_-_120331936 | 0.75 |
ENSMUST00000093099.13
|
Taf1c
|
TATA-box binding protein associated factor, RNA polymerase I, C |
| chr7_-_126626152 | 0.75 |
ENSMUST00000206254.2
ENSMUST00000206291.2 |
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr3_+_96604390 | 0.75 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr12_+_12312163 | 0.74 |
ENSMUST00000069005.10
ENSMUST00000223061.2 |
Cyria
|
CYFIP related Rac1 interactor A |
| chr10_+_126814542 | 0.74 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr19_+_42135812 | 0.74 |
ENSMUST00000061111.10
|
Marveld1
|
MARVEL (membrane-associating) domain containing 1 |
| chr6_-_3487369 | 0.74 |
ENSMUST00000201607.4
|
Hepacam2
|
HEPACAM family member 2 |
| chr14_+_30853010 | 0.74 |
ENSMUST00000227096.2
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr5_-_137101108 | 0.73 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr7_+_3337591 | 0.73 |
ENSMUST00000203328.4
|
Myadm
|
myeloid-associated differentiation marker |
| chr17_+_29251602 | 0.72 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr11_+_69737491 | 0.72 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr15_-_79287447 | 0.71 |
ENSMUST00000228002.2
|
Tmem184b
|
transmembrane protein 184b |
| chr19_+_6952580 | 0.70 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr4_+_62204678 | 0.70 |
ENSMUST00000084530.9
|
Slc31a2
|
solute carrier family 31, member 2 |
| chr9_+_65816206 | 0.70 |
ENSMUST00000205379.2
ENSMUST00000206048.2 ENSMUST00000034949.10 ENSMUST00000154589.2 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr16_+_33504908 | 0.69 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr5_-_107437427 | 0.69 |
ENSMUST00000031224.15
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr1_+_171216480 | 0.69 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr9_+_71123061 | 0.68 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr17_-_66191912 | 0.68 |
ENSMUST00000024905.11
|
Ralbp1
|
ralA binding protein 1 |
| chr7_-_66339319 | 0.68 |
ENSMUST00000124899.8
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr1_-_106641940 | 0.68 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr15_-_83395065 | 0.67 |
ENSMUST00000109479.8
ENSMUST00000109480.8 ENSMUST00000016897.12 |
Ttll1
|
tubulin tyrosine ligase-like 1 |
| chr6_+_34453142 | 0.66 |
ENSMUST00000045372.6
ENSMUST00000138668.2 ENSMUST00000139067.2 |
Bpgm
|
2,3-bisphosphoglycerate mutase |
| chr11_+_11634967 | 0.66 |
ENSMUST00000141436.8
ENSMUST00000126058.8 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr8_+_85449632 | 0.66 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr7_+_126359763 | 0.65 |
ENSMUST00000091328.4
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr19_+_38919477 | 0.65 |
ENSMUST00000145051.2
|
Hells
|
helicase, lymphoid specific |
| chr6_-_51446752 | 0.65 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr1_-_87029312 | 0.65 |
ENSMUST00000113270.3
|
Alpi
|
alkaline phosphatase, intestinal |
| chr11_+_96355441 | 0.65 |
ENSMUST00000071510.14
ENSMUST00000107662.9 |
Skap1
|
src family associated phosphoprotein 1 |
| chr4_-_33248499 | 0.65 |
ENSMUST00000049357.10
|
Pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_+_27750349 | 0.64 |
ENSMUST00000033880.7
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr16_+_32427738 | 0.64 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr5_+_17779273 | 0.64 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr11_+_69737437 | 0.64 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr7_+_66339637 | 0.63 |
ENSMUST00000153007.2
ENSMUST00000121777.9 ENSMUST00000150071.8 ENSMUST00000077967.13 |
Lins1
|
lines homolog 1 |
| chr7_+_141047298 | 0.63 |
ENSMUST00000106000.10
ENSMUST00000209892.2 ENSMUST00000177840.9 |
Cd151
|
CD151 antigen |
| chr17_+_36134122 | 0.63 |
ENSMUST00000001569.15
ENSMUST00000174080.8 |
Flot1
|
flotillin 1 |
| chr5_+_91287448 | 0.62 |
ENSMUST00000031325.6
|
Areg
|
amphiregulin |
| chr9_+_54606832 | 0.61 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr5_+_137628377 | 0.61 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr12_-_112640626 | 0.60 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr4_-_133695204 | 0.60 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr17_-_27247581 | 0.60 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr9_-_107749982 | 0.59 |
ENSMUST00000183032.8
ENSMUST00000035201.13 |
Rbm6
|
RNA binding motif protein 6 |
| chr11_-_100830183 | 0.59 |
ENSMUST00000092671.12
ENSMUST00000103114.8 |
Stat3
|
signal transducer and activator of transcription 3 |
| chr4_+_136038243 | 0.59 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr8_-_8711211 | 0.58 |
ENSMUST00000001319.15
|
Efnb2
|
ephrin B2 |
| chr4_-_135601098 | 0.58 |
ENSMUST00000030436.12
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr12_+_12312135 | 0.58 |
ENSMUST00000069066.14
|
Cyria
|
CYFIP related Rac1 interactor A |
| chr3_+_87837566 | 0.57 |
ENSMUST00000055984.7
|
Isg20l2
|
interferon stimulated exonuclease gene 20-like 2 |
| chr3_-_94566107 | 0.57 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr19_+_5540591 | 0.57 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle |
| chr2_+_29968596 | 0.57 |
ENSMUST00000045246.8
|
Pkn3
|
protein kinase N3 |
| chr18_+_44237577 | 0.57 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr17_+_36134398 | 0.56 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr13_-_114595122 | 0.56 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr5_+_73071695 | 0.56 |
ENSMUST00000143829.5
|
Slain2
|
SLAIN motif family, member 2 |
| chr12_-_100691316 | 0.56 |
ENSMUST00000222731.2
|
Rps6ka5
|
ribosomal protein S6 kinase, polypeptide 5 |
| chrX_-_37341274 | 0.56 |
ENSMUST00000089056.10
ENSMUST00000089054.11 ENSMUST00000066498.8 |
Tmem255a
|
transmembrane protein 255A |
| chr18_+_44237474 | 0.55 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr17_+_9207180 | 0.55 |
ENSMUST00000151609.2
ENSMUST00000232775.2 ENSMUST00000136954.3 |
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr19_+_21249636 | 0.55 |
ENSMUST00000237651.2
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr3_-_87837403 | 0.55 |
ENSMUST00000005016.16
|
Rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr8_+_96058907 | 0.55 |
ENSMUST00000034245.16
|
Usb1
|
U6 snRNA biogenesis 1 |
| chr11_-_87249837 | 0.54 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr8_-_57003828 | 0.54 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr18_+_11052458 | 0.54 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr10_-_79938487 | 0.54 |
ENSMUST00000042771.8
|
Sbno2
|
strawberry notch 2 |
| chr13_-_101904662 | 0.54 |
ENSMUST00000055518.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr9_+_54606144 | 0.53 |
ENSMUST00000120452.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 1.7 | 5.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.8 | 3.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.8 | 3.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 2.8 | GO:0051673 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 5.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.6 | 2.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.6 | 9.0 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.5 | 2.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.5 | 1.6 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.5 | 1.9 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.4 | 2.6 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.4 | 1.5 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.4 | 1.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.4 | 2.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 1.7 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.3 | 1.0 | GO:1902867 | negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.3 | 1.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 1.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.3 | 0.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.3 | 2.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.3 | 1.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 1.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 0.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 1.0 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 0.9 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 1.9 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 0.9 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.2 | 3.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 2.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 1.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 0.7 | GO:0060936 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.2 | 0.7 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.2 | 0.7 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.2 | 1.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 1.5 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 0.6 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 3.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 1.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.6 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.2 | 2.0 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.9 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.2 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.5 | GO:1900238 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 | 0.5 | GO:0097037 | heme export(GO:0097037) |
| 0.2 | 1.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0072076 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 1.9 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 1.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 1.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.5 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 2.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 1.5 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.6 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.1 | 0.7 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.1 | 1.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 1.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.3 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 2.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.6 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.7 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 1.8 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.3 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.9 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.7 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 2.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.6 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 1.0 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 3.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.8 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.9 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 1.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 1.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.0 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.9 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 1.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 3.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 | 0.4 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.6 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 1.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 1.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 2.0 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.3 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.0 | 1.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 2.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 2.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.7 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.5 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 3.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.2 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 2.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 1.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 1.3 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 1.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.3 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.5 | GO:0000910 | cytokinesis(GO:0000910) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.9 | 2.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.9 | 5.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 2.8 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 3.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 5.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 1.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 0.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 2.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.6 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 3.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 3.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 6.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 4.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 9.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.9 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 3.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.3 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.7 | 2.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.6 | 12.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 1.6 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.5 | 9.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.5 | 3.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 1.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.4 | 2.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 3.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 3.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.3 | 2.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 0.9 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 1.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 10.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.8 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.2 | 3.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 3.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 5.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 1.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 5.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.3 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 4.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 4.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 4.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.7 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 2.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 4.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 2.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 1.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 2.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 2.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 6.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 7.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 7.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 9.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 5.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 2.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 8.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 6.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 3.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 7.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 4.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |