Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
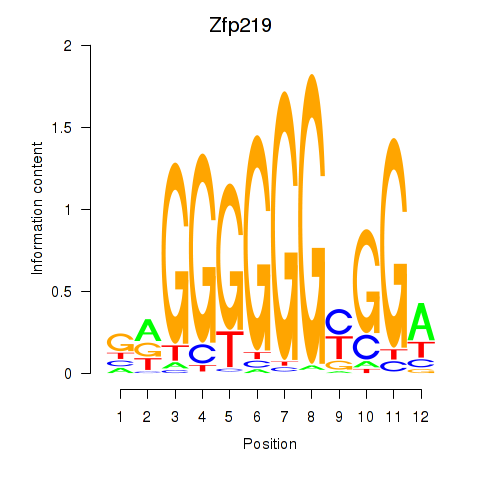
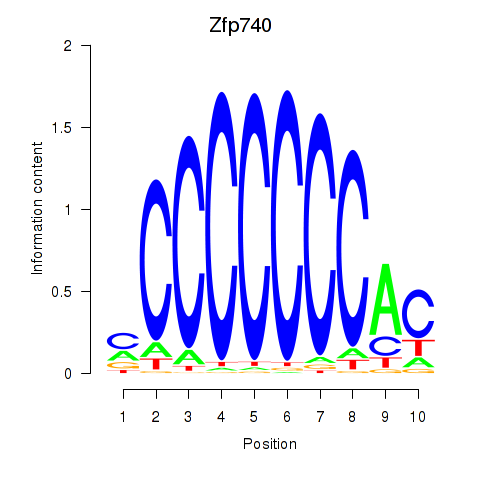
Results for Zfp219_Zfp740
Z-value: 1.50
Transcription factors associated with Zfp219_Zfp740
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp219
|
ENSMUSG00000049295.18 | Zfp219 |
|
Zfp740
|
ENSMUSG00000046897.18 | Zfp740 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp740 | mm39_v1_chr15_+_102112074_102112144 | -0.11 | 3.6e-01 | Click! |
| Zfp219 | mm39_v1_chr14_-_52252318_52252369 | -0.10 | 4.1e-01 | Click! |
Activity profile of Zfp219_Zfp740 motif
Sorted Z-values of Zfp219_Zfp740 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp219_Zfp740
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126625739 | 24.96 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_-_126625657 | 16.42 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_-_88609048 | 13.72 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_-_126625617 | 13.58 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_-_88608958 | 13.21 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr11_+_96242422 | 11.06 |
ENSMUST00000100523.7
|
Hoxb2
|
homeobox B2 |
| chr11_+_96173475 | 10.86 |
ENSMUST00000168043.2
|
Hoxb8
|
homeobox B8 |
| chr11_+_117740077 | 10.71 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr11_+_96173355 | 10.33 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chr7_-_126626152 | 10.19 |
ENSMUST00000206254.2
ENSMUST00000206291.2 |
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr4_-_11386756 | 9.60 |
ENSMUST00000108313.8
ENSMUST00000108311.9 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr17_+_34808772 | 9.12 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr2_-_169973076 | 9.03 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr4_-_11386679 | 8.73 |
ENSMUST00000043781.14
ENSMUST00000108310.8 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr11_+_96162283 | 8.60 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr15_+_102314809 | 8.57 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr18_+_64473091 | 8.52 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr4_-_133746138 | 7.38 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr17_+_34809132 | 7.30 |
ENSMUST00000173772.2
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr13_+_55359598 | 7.02 |
ENSMUST00000224918.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr4_-_3938352 | 6.81 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr9_-_110571645 | 6.66 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr2_+_78699360 | 6.35 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_-_89300599 | 6.30 |
ENSMUST00000142119.2
ENSMUST00000029677.9 ENSMUST00000148361.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr3_-_89300936 | 6.23 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr3_+_88439616 | 6.22 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr4_+_41135730 | 6.18 |
ENSMUST00000040008.4
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr1_+_74430575 | 6.16 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr18_+_34910064 | 6.11 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chrX_-_37653396 | 6.08 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr5_+_53748323 | 6.04 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr17_+_35113490 | 6.03 |
ENSMUST00000052778.10
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr2_-_38604503 | 6.02 |
ENSMUST00000028084.5
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr18_+_82929451 | 6.00 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr2_+_156681991 | 5.95 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr2_+_4886298 | 5.90 |
ENSMUST00000027973.14
|
Sephs1
|
selenophosphate synthetase 1 |
| chr12_-_118265163 | 5.79 |
ENSMUST00000221844.2
|
Sp4
|
trans-acting transcription factor 4 |
| chr11_+_96177449 | 5.76 |
ENSMUST00000049352.8
|
Hoxb7
|
homeobox B7 |
| chr4_+_3938881 | 5.75 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_+_130904000 | 5.57 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr19_-_45731312 | 5.46 |
ENSMUST00000026241.12
ENSMUST00000026240.14 ENSMUST00000111928.8 |
Fgf8
|
fibroblast growth factor 8 |
| chr17_+_34811217 | 5.27 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr2_-_147887810 | 5.07 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr3_+_41519289 | 5.04 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr2_+_4887015 | 5.03 |
ENSMUST00000115019.2
|
Sephs1
|
selenophosphate synthetase 1 |
| chr11_+_95603494 | 4.85 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr19_+_4264292 | 4.83 |
ENSMUST00000046506.7
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chrX_+_41238410 | 4.81 |
ENSMUST00000127618.8
|
Stag2
|
stromal antigen 2 |
| chr15_+_102314578 | 4.76 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chr12_-_118265103 | 4.74 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr6_+_120643323 | 4.69 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr4_-_11386394 | 4.63 |
ENSMUST00000155519.2
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr11_+_23256883 | 4.61 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_+_77873535 | 4.46 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr19_+_37423198 | 4.44 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr6_+_124986224 | 4.41 |
ENSMUST00000112427.8
|
Zfp384
|
zinc finger protein 384 |
| chr5_+_115644727 | 4.40 |
ENSMUST00000067268.15
ENSMUST00000086523.7 ENSMUST00000212819.3 |
Pxn
|
paxillin |
| chr6_+_124986078 | 4.38 |
ENSMUST00000054553.11
|
Zfp384
|
zinc finger protein 384 |
| chr11_+_3239868 | 4.29 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr6_+_124986627 | 4.23 |
ENSMUST00000046064.17
ENSMUST00000152752.8 ENSMUST00000088308.10 ENSMUST00000112425.8 ENSMUST00000084275.12 |
Zfp384
|
zinc finger protein 384 |
| chr6_+_124986193 | 4.23 |
ENSMUST00000112428.8
|
Zfp384
|
zinc finger protein 384 |
| chr18_+_82929037 | 4.17 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr10_-_7831657 | 4.13 |
ENSMUST00000147938.2
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr7_+_44114815 | 4.10 |
ENSMUST00000035929.11
ENSMUST00000146128.8 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr1_-_84817000 | 4.07 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr9_+_100525807 | 4.02 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1 |
| chr7_-_89629809 | 3.91 |
ENSMUST00000238792.2
|
Eed
|
embryonic ectoderm development |
| chr13_-_30168374 | 3.90 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr5_+_123280250 | 3.85 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr10_-_4338032 | 3.82 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr4_-_87951565 | 3.76 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr7_+_44114857 | 3.73 |
ENSMUST00000135624.2
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr2_-_84605732 | 3.69 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr18_+_82928959 | 3.65 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr2_+_174171799 | 3.63 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr4_+_65042411 | 3.63 |
ENSMUST00000084501.4
|
Pappa
|
pregnancy-associated plasma protein A |
| chr11_+_77405933 | 3.53 |
ENSMUST00000094004.5
|
Abhd15
|
abhydrolase domain containing 15 |
| chrX_+_70600481 | 3.46 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr18_+_35963353 | 3.40 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr15_+_102379621 | 3.37 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr19_-_3736549 | 3.29 |
ENSMUST00000025856.17
ENSMUST00000176867.2 |
Lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr2_+_153334710 | 3.25 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr17_+_34134873 | 3.24 |
ENSMUST00000172619.8
ENSMUST00000174463.2 |
Tapbp
Zbtb22
|
TAP binding protein zinc finger and BTB domain containing 22 |
| chr15_+_102379503 | 3.22 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr15_+_96185399 | 3.20 |
ENSMUST00000134985.9
ENSMUST00000096250.5 |
Arid2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr10_-_7831979 | 3.17 |
ENSMUST00000146444.8
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr11_-_102208615 | 3.17 |
ENSMUST00000107117.9
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr2_+_31530049 | 3.16 |
ENSMUST00000113470.3
|
Prdm12
|
PR domain containing 12 |
| chr5_-_123126550 | 3.15 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr2_+_152873772 | 3.15 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr17_+_47905553 | 3.14 |
ENSMUST00000182846.3
|
Ccnd3
|
cyclin D3 |
| chr18_+_35962832 | 3.12 |
ENSMUST00000060722.8
|
Cxxc5
|
CXXC finger 5 |
| chr7_+_89779564 | 3.08 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_+_127084283 | 3.08 |
ENSMUST00000048896.8
|
Fbrs
|
fibrosin |
| chr10_+_43455157 | 3.07 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr18_+_56840813 | 3.07 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr3_+_106389732 | 3.06 |
ENSMUST00000029508.11
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr2_+_27055245 | 3.05 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr1_+_133291302 | 3.05 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr17_-_28569721 | 3.04 |
ENSMUST00000156862.3
|
Tead3
|
TEA domain family member 3 |
| chr9_+_72439891 | 3.03 |
ENSMUST00000183372.8
ENSMUST00000184015.2 |
Rfx7
|
regulatory factor X, 7 |
| chr6_-_52237765 | 3.02 |
ENSMUST00000147595.7
|
Hoxa13
|
homeobox A13 |
| chr6_-_72212547 | 3.01 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr6_-_47571901 | 3.00 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr15_+_81469538 | 2.99 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr4_-_129083335 | 2.99 |
ENSMUST00000106061.9
ENSMUST00000072431.13 |
S100pbp
|
S100P binding protein |
| chr5_-_34345014 | 2.98 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr2_+_127967951 | 2.97 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr2_+_156681927 | 2.95 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr10_+_127478844 | 2.95 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr7_-_89630141 | 2.93 |
ENSMUST00000238981.2
ENSMUST00000208977.2 ENSMUST00000107234.3 |
Eed
|
embryonic ectoderm development |
| chr11_+_98851238 | 2.93 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr19_+_6952580 | 2.90 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr11_-_78056347 | 2.90 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr11_+_96194333 | 2.88 |
ENSMUST00000049272.5
|
Hoxb5
|
homeobox B5 |
| chr4_-_59549243 | 2.88 |
ENSMUST00000173699.8
ENSMUST00000173884.8 ENSMUST00000102883.11 ENSMUST00000174586.8 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr4_-_129083439 | 2.87 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr6_-_125213911 | 2.86 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr9_+_61280501 | 2.86 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr4_+_108317197 | 2.84 |
ENSMUST00000097925.9
|
Tut4
|
terminal uridylyl transferase 4 |
| chr4_-_129083251 | 2.82 |
ENSMUST00000117965.8
|
S100pbp
|
S100P binding protein |
| chr4_-_133480922 | 2.82 |
ENSMUST00000145664.9
ENSMUST00000105897.10 |
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr14_-_52252003 | 2.81 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr10_-_127502541 | 2.81 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chrX_-_47543029 | 2.78 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chrX_+_41238193 | 2.76 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr5_-_147244074 | 2.74 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr11_+_97554192 | 2.73 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr4_-_43523745 | 2.72 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr15_+_102875229 | 2.71 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr7_+_127376550 | 2.71 |
ENSMUST00000126761.8
ENSMUST00000047157.13 |
Setd1a
|
SET domain containing 1A |
| chr2_+_103800553 | 2.71 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr19_-_45731290 | 2.69 |
ENSMUST00000111927.8
|
Fgf8
|
fibroblast growth factor 8 |
| chr9_+_62765362 | 2.68 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr17_-_71158184 | 2.64 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr17_-_71158052 | 2.64 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr14_-_55828511 | 2.62 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr12_-_32000169 | 2.61 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr11_-_102076028 | 2.60 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr15_-_102112159 | 2.58 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr2_+_75489596 | 2.58 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr12_+_87193922 | 2.57 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr5_+_97145533 | 2.57 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr10_+_62860094 | 2.56 |
ENSMUST00000124784.8
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr11_-_75330302 | 2.54 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr1_-_191129223 | 2.53 |
ENSMUST00000067976.9
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr11_-_106811507 | 2.53 |
ENSMUST00000103067.10
|
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr5_-_134485081 | 2.52 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr11_-_70545450 | 2.52 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chr11_-_75330415 | 2.51 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr17_-_28569574 | 2.51 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr10_+_127928622 | 2.49 |
ENSMUST00000219072.2
ENSMUST00000045621.9 ENSMUST00000170054.9 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr5_-_143717970 | 2.48 |
ENSMUST00000053287.6
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr15_+_102927366 | 2.48 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr5_-_107873883 | 2.48 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr1_+_172328768 | 2.46 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr5_-_142891686 | 2.45 |
ENSMUST00000106216.3
|
Actb
|
actin, beta |
| chr11_-_69812053 | 2.44 |
ENSMUST00000108613.10
ENSMUST00000043419.10 ENSMUST00000070996.11 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr2_+_174171860 | 2.44 |
ENSMUST00000109087.8
ENSMUST00000109084.8 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr3_-_89245297 | 2.43 |
ENSMUST00000029674.8
|
Efna4
|
ephrin A4 |
| chr16_-_10603389 | 2.43 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr19_+_6952319 | 2.37 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr15_-_50753437 | 2.37 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr4_+_8691303 | 2.35 |
ENSMUST00000051558.10
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr7_-_44180700 | 2.34 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr5_-_137529465 | 2.34 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr8_+_84724130 | 2.34 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr11_-_88608920 | 2.33 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr10_-_37014859 | 2.30 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr4_-_136563154 | 2.29 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr19_+_44745833 | 2.28 |
ENSMUST00000004340.11
|
Pax2
|
paired box 2 |
| chr10_-_127502467 | 2.28 |
ENSMUST00000099157.4
|
Nab2
|
Ngfi-A binding protein 2 |
| chr11_+_117157024 | 2.27 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr19_+_55730316 | 2.27 |
ENSMUST00000111658.10
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr10_+_79690452 | 2.26 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr4_-_43523595 | 2.25 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr15_+_102427149 | 2.25 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr10_-_127124718 | 2.24 |
ENSMUST00000156208.2
ENSMUST00000026476.13 |
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr10_-_93146937 | 2.24 |
ENSMUST00000008542.12
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr2_+_109111083 | 2.21 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr7_-_126736979 | 2.21 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr11_-_70545424 | 2.19 |
ENSMUST00000108549.2
|
Pfn1
|
profilin 1 |
| chr2_-_34803988 | 2.19 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr17_+_85928459 | 2.18 |
ENSMUST00000162695.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr12_-_32000209 | 2.18 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr10_+_79986280 | 2.17 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chr13_+_111822759 | 2.17 |
ENSMUST00000231979.2
|
Mier3
|
MIER family member 3 |
| chrX_-_140508177 | 2.17 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr10_-_24803336 | 2.16 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr5_-_123127346 | 2.15 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_-_115109118 | 2.12 |
ENSMUST00000031535.12
|
Hnf1a
|
HNF1 homeobox A |
| chr18_+_73706115 | 2.12 |
ENSMUST00000091852.5
|
Mex3c
|
mex3 RNA binding family member C |
| chr1_-_180641099 | 2.11 |
ENSMUST00000159789.2
ENSMUST00000081026.11 |
H3f3a
|
H3.3 histone A |
| chr17_-_32607859 | 2.11 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr18_+_35962881 | 2.10 |
ENSMUST00000237291.2
|
Cxxc5
|
CXXC finger 5 |
| chr4_-_129083392 | 2.10 |
ENSMUST00000117497.8
ENSMUST00000117350.2 |
S100pbp
|
S100P binding protein |
| chr1_-_180641159 | 2.09 |
ENSMUST00000162118.8
ENSMUST00000159685.2 ENSMUST00000161308.8 |
H3f3a
|
H3.3 histone A |
| chr15_-_8473918 | 2.07 |
ENSMUST00000052965.8
|
Nipbl
|
NIPBL cohesin loading factor |
| chr2_-_179618439 | 2.07 |
ENSMUST00000041618.13
ENSMUST00000227325.2 |
Taf4
|
TATA-box binding protein associated factor 4 |
| chr9_+_100956145 | 2.06 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 65.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 4.4 | 13.3 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 4.1 | 12.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 3.1 | 12.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 2.3 | 6.8 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 1.8 | 9.2 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 1.8 | 7.0 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 1.7 | 5.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.4 | 8.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 1.4 | 1.4 | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 1.4 | 5.6 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 1.3 | 10.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 1.3 | 7.8 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.3 | 3.9 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 1.3 | 7.8 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 1.3 | 9.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 1.2 | 7.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.2 | 3.7 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.2 | 6.1 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 1.1 | 10.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.1 | 3.2 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 1.0 | 3.1 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.0 | 3.0 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.0 | 6.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 1.0 | 6.0 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 1.0 | 3.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 1.0 | 8.9 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 1.0 | 2.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.0 | 4.9 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 1.0 | 5.8 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 1.0 | 3.9 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.9 | 3.8 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.9 | 2.8 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.9 | 6.5 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.9 | 2.7 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.9 | 3.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.9 | 3.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.9 | 3.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.9 | 26.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.8 | 2.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.8 | 3.1 | GO:1904008 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.8 | 6.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.8 | 2.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.8 | 2.3 | GO:2000597 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.7 | 16.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.7 | 3.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.7 | 2.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.7 | 6.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.7 | 1.4 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.7 | 4.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) |
| 0.7 | 2.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.7 | 4.8 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.6 | 3.8 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 2.5 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.6 | 1.8 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.6 | 3.0 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.6 | 16.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.6 | 12.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.6 | 3.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.6 | 2.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.6 | 5.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.6 | 2.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.6 | 1.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.6 | 5.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 2.2 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.5 | 2.7 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.5 | 3.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.5 | 3.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.5 | 2.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.5 | 3.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.5 | 0.5 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.5 | 23.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.5 | 4.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.5 | 1.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.5 | 4.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 5.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.5 | 5.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.4 | 8.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 1.8 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.4 | 1.8 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.4 | 1.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 1.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 2.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.4 | 3.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 1.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.4 | 1.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 2.8 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.4 | 2.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.4 | 3.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.4 | 4.7 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.4 | 1.2 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.4 | 1.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 3.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 2.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) negative regulation of defense response to virus by host(GO:0050689) |
| 0.4 | 5.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.4 | 0.7 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 5.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.4 | 1.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 0.7 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.4 | 3.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 2.4 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.3 | 2.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.3 | 2.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 1.0 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.3 | 1.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 1.6 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 1.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 6.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.3 | 0.9 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.3 | 4.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 2.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 1.5 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.3 | 0.9 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.3 | 2.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 17.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.3 | 3.2 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.3 | 2.6 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.3 | 3.5 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.3 | 5.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 1.1 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.3 | 2.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.3 | 6.8 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 1.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 | 2.8 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 4.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 2.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.3 | 0.8 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.3 | 4.8 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.3 | 0.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.3 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 1.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.3 | 1.8 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.3 | 1.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 2.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 0.5 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.3 | 4.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.3 | 1.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 2.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 3.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 1.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 0.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 1.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 0.9 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.9 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 2.0 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.2 | 0.9 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 1.7 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 2.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.6 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.2 | 0.6 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.2 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 4.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 5.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 2.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 0.7 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 1.5 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.2 | 2.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.5 | GO:0036090 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.2 | 2.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.0 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 | 1.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 2.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 2.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 6.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 2.9 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 1.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 1.5 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.2 | 0.8 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 1.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 4.4 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.6 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.6 | GO:0001805 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) positive regulation of mast cell cytokine production(GO:0032765) negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.1 | 2.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.8 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 1.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.1 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.1 | 3.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 2.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.7 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.1 | 0.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.7 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 1.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.6 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 | 1.6 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 1.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.1 | GO:0021938 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 4.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 23.2 | GO:0048864 | stem cell development(GO:0048864) |
| 0.1 | 4.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 3.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 10.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 2.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.8 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.1 | 0.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 1.2 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 1.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.2 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 1.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 5.7 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 2.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 6.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 5.3 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.4 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.1 | 1.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.5 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 1.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 1.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.1 | 1.0 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 8.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.2 | GO:2000722 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 4.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 1.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 1.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.9 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 1.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.0 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 1.8 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) small RNA loading onto RISC(GO:0070922) |
| 0.1 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 1.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 2.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.8 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 1.6 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 0.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 3.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.9 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 1.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 1.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.9 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 3.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 13.1 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.0 | 0.9 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 2.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 2.2 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.6 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 1.0 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 1.0 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 2.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 2.5 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 0.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 3.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 6.0 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.1 | GO:0001656 | metanephros development(GO:0001656) |
| 0.0 | 0.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 6.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.5 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.0 | 0.2 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 4.9 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 3.1 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.3 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0060903 | regulation of meiosis I(GO:0060631) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.5 | 6.1 | GO:0001740 | Barr body(GO:0001740) |
| 1.0 | 3.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.9 | 12.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.9 | 2.6 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.9 | 6.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.9 | 6.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.8 | 9.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.8 | 3.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.8 | 6.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.7 | 5.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.7 | 6.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 7.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.6 | 1.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.6 | 1.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.5 | 7.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 2.4 | GO:0035363 | histone locus body(GO:0035363) |
| 0.5 | 3.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 5.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.4 | 3.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 3.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 1.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 2.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 28.3 | GO:0005844 | polysome(GO:0005844) |
| 0.3 | 1.9 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.3 | 2.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 2.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.3 | 2.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 1.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.3 | 4.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 7.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 1.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 5.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 2.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 8.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.2 | 21.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 1.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 1.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.6 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 1.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 0.6 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.2 | 1.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 1.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.8 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 9.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 20.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 2.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 3.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 3.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 56.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 3.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 26.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.5 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 3.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 4.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 66.8 | GO:0044451 | nucleoplasm part(GO:0044451) |
| 0.1 | 2.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 3.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 4.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 6.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 30.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 3.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 9.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 53.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 5.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 60.9 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 1.6 | 8.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.4 | 4.2 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 1.3 | 5.2 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 1.3 | 12.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.3 | 8.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.1 | 3.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.0 | 3.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.0 | 3.9 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.0 | 6.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.9 | 7.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.9 | 29.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.8 | 4.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.8 | 2.4 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.8 | 5.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.8 | 108.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.8 | 10.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.8 | 3.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.7 | 6.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.6 | 2.6 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 1.6 | GO:0071820 | N-box binding(GO:0071820) |
| 0.5 | 7.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.5 | 2.7 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.5 | 2.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.5 | 3.5 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.5 | 8.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.5 | 1.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 1.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 1.4 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.4 | 1.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 1.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 2.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 4.7 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 4.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 9.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 3.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 3.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 3.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 1.5 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 1.9 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 2.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 1.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 5.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 2.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 7.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 2.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 5.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 4.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 1.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 2.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 2.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.3 | 6.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 4.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 3.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 4.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 4.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 12.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.3 | 0.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 14.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 7.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 4.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 13.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.3 | 1.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 1.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 5.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 2.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 1.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 2.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.9 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 1.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 5.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 2.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 6.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.2 | 2.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 9.5 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 1.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 2.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 6.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 1.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 6.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 4.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 1.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 4.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 1.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 2.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 1.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 101.0 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.2 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 1.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 35.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.6 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.1 | 0.8 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 1.1 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 3.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 6.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 73.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.1 | 8.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 2.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 2.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 4.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 7.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 3.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 2.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.8 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 20.7 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 10.8 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 6.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 4.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 28.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.4 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 1.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.3 | 16.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 11.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 13.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 14.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 9.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 5.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 8.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 7.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 13.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 7.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 6.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 6.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 13.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.7 | 29.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.7 | 11.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 14.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.4 | 8.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 21.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 8.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 8.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 9.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 7.8 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.2 | 2.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.2 | 3.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 6.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 8.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 1.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 15.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 6.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 7.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 5.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 5.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 2.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 9.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 0.7 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 3.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.1 | 23.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 10.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 6.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 5.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 3.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 5.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 6.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |