Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
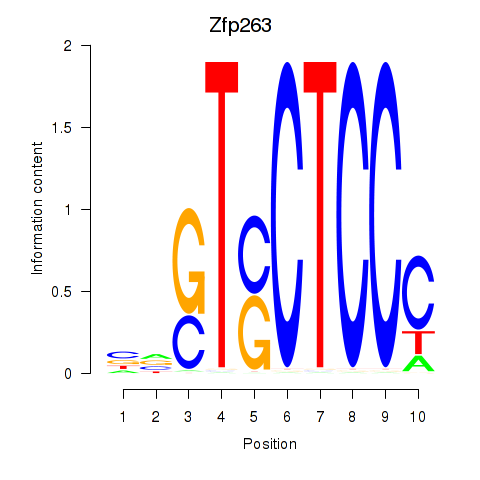
Results for Zfp263
Z-value: 1.19
Transcription factors associated with Zfp263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp263
|
ENSMUSG00000022529.12 | Zfp263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | mm39_v1_chr16_+_3561952_3562023 | 0.05 | 6.8e-01 | Click! |
Activity profile of Zfp263 motif
Sorted Z-values of Zfp263 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp263
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_26028059 | 7.79 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chrX_+_135723531 | 5.81 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr3_+_83673606 | 5.72 |
ENSMUST00000029625.8
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr9_-_83688294 | 5.41 |
ENSMUST00000034796.14
ENSMUST00000183614.2 |
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 |
| chr19_-_58849407 | 5.23 |
ENSMUST00000066285.6
|
Hspa12a
|
heat shock protein 12A |
| chr14_-_24053994 | 5.05 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_+_135723420 | 4.97 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr9_+_66621001 | 4.92 |
ENSMUST00000085420.12
|
Car12
|
carbonic anhydrase 12 |
| chr9_+_66620959 | 4.91 |
ENSMUST00000071889.13
|
Car12
|
carbonic anhydrase 12 |
| chr1_-_172125555 | 4.74 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr7_-_127423641 | 4.40 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B |
| chr2_+_156263002 | 4.40 |
ENSMUST00000125153.10
ENSMUST00000103136.8 ENSMUST00000109577.9 |
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_+_105183463 | 4.23 |
ENSMUST00000100335.10
ENSMUST00000021038.5 |
Mrc2
|
mannose receptor, C type 2 |
| chr7_-_30826184 | 4.06 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr18_+_34994253 | 4.04 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr14_-_55163311 | 4.00 |
ENSMUST00000022813.8
|
Efs
|
embryonal Fyn-associated substrate |
| chr1_-_17168063 | 3.87 |
ENSMUST00000038382.5
|
Jph1
|
junctophilin 1 |
| chr5_-_39801940 | 3.85 |
ENSMUST00000152057.2
ENSMUST00000053116.7 |
Hs3st1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr2_+_145009625 | 3.78 |
ENSMUST00000110007.8
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr11_+_5811886 | 3.72 |
ENSMUST00000102923.10
|
Aebp1
|
AE binding protein 1 |
| chr1_-_168259710 | 3.66 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr14_-_55354392 | 3.63 |
ENSMUST00000022819.13
|
Jph4
|
junctophilin 4 |
| chr6_-_124840824 | 3.61 |
ENSMUST00000046893.10
ENSMUST00000204667.2 |
Gpr162
|
G protein-coupled receptor 162 |
| chr3_+_90444537 | 3.54 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr11_+_17001818 | 3.54 |
ENSMUST00000058159.6
|
Cnrip1
|
cannabinoid receptor interacting protein 1 |
| chr1_-_170417354 | 3.47 |
ENSMUST00000160456.8
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr16_+_7011580 | 3.35 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_+_98632696 | 3.34 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr3_+_90444613 | 3.33 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr1_+_34840785 | 3.32 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr4_-_43523388 | 3.31 |
ENSMUST00000107913.10
ENSMUST00000030184.12 |
Tpm2
|
tropomyosin 2, beta |
| chr2_+_156262957 | 3.26 |
ENSMUST00000109574.8
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_+_98632953 | 3.25 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr15_-_75963446 | 3.13 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr14_-_24054927 | 3.10 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_+_109760856 | 3.09 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr11_-_46057224 | 3.05 |
ENSMUST00000020679.3
|
Nipal4
|
NIPA-like domain containing 4 |
| chr10_+_80765900 | 2.98 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_+_98632631 | 2.97 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr3_+_157272504 | 2.94 |
ENSMUST00000041175.13
ENSMUST00000173533.2 |
Ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr14_-_55163452 | 2.94 |
ENSMUST00000227037.2
|
Efs
|
embryonal Fyn-associated substrate |
| chr15_+_98006346 | 2.92 |
ENSMUST00000051226.8
|
Pfkm
|
phosphofructokinase, muscle |
| chr1_+_163607143 | 2.91 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr5_-_91550853 | 2.90 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr1_+_172383499 | 2.89 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr2_+_156262756 | 2.88 |
ENSMUST00000103137.10
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr17_-_90763300 | 2.87 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr4_-_126647156 | 2.86 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chr12_-_76842263 | 2.82 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr17_+_35455532 | 2.80 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr17_+_34879431 | 2.79 |
ENSMUST00000238967.2
|
Tnxb
|
tenascin XB |
| chr9_-_54554483 | 2.79 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_-_115894993 | 2.78 |
ENSMUST00000074285.8
|
Meis2
|
Meis homeobox 2 |
| chr2_+_35146390 | 2.77 |
ENSMUST00000201185.4
ENSMUST00000202990.4 ENSMUST00000202899.4 ENSMUST00000142324.8 ENSMUST00000139867.5 |
Gsn
|
gelsolin |
| chr6_-_53797748 | 2.74 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr7_-_105131407 | 2.73 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr2_+_25132941 | 2.71 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr19_+_28812474 | 2.69 |
ENSMUST00000025875.5
|
Slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr18_+_36475573 | 2.68 |
ENSMUST00000139727.3
ENSMUST00000237375.2 ENSMUST00000235403.2 ENSMUST00000236593.2 ENSMUST00000236374.2 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr13_+_46655589 | 2.67 |
ENSMUST00000119341.2
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr3_+_96503944 | 2.66 |
ENSMUST00000058943.8
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr17_-_56440817 | 2.65 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr9_-_44632680 | 2.65 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr3_+_93227047 | 2.63 |
ENSMUST00000090856.10
ENSMUST00000093774.4 |
Hrnr
|
hornerin |
| chr11_-_3321307 | 2.60 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr13_+_19132375 | 2.59 |
ENSMUST00000239207.2
ENSMUST00000003345.10 ENSMUST00000200466.5 |
Amph
|
amphiphysin |
| chr3_+_54063459 | 2.58 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr3_+_75464837 | 2.58 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr8_+_31581635 | 2.58 |
ENSMUST00000161713.2
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr17_+_81251997 | 2.58 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr11_+_101221431 | 2.56 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr7_+_29007349 | 2.55 |
ENSMUST00000108230.8
ENSMUST00000065181.12 |
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr11_+_17001586 | 2.55 |
ENSMUST00000131515.2
|
Cnrip1
|
cannabinoid receptor interacting protein 1 |
| chr18_-_43032535 | 2.55 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr14_+_68321302 | 2.54 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr19_-_58849380 | 2.54 |
ENSMUST00000235263.2
|
Hspa12a
|
heat shock protein 12A |
| chr16_+_20551853 | 2.53 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr2_-_115895528 | 2.52 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
| chr11_+_35011953 | 2.51 |
ENSMUST00000069837.4
|
Slit3
|
slit guidance ligand 3 |
| chr9_+_23134372 | 2.50 |
ENSMUST00000071982.7
|
Bmper
|
BMP-binding endothelial regulator |
| chr9_-_37058590 | 2.48 |
ENSMUST00000080754.12
ENSMUST00000188057.7 ENSMUST00000039674.13 |
Pknox2
|
Pbx/knotted 1 homeobox 2 |
| chr6_-_114018982 | 2.46 |
ENSMUST00000101045.10
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr18_-_12995681 | 2.42 |
ENSMUST00000121808.8
ENSMUST00000118313.8 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr7_+_73025243 | 2.41 |
ENSMUST00000119206.3
ENSMUST00000094312.12 |
Rgma
|
repulsive guidance molecule family member A |
| chr12_+_37930661 | 2.40 |
ENSMUST00000040500.9
|
Dgkb
|
diacylglycerol kinase, beta |
| chr2_-_115895202 | 2.40 |
ENSMUST00000110906.9
|
Meis2
|
Meis homeobox 2 |
| chr17_-_14914484 | 2.37 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr3_-_108133914 | 2.37 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr18_-_12995261 | 2.36 |
ENSMUST00000234427.2
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr13_+_46655324 | 2.32 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr6_-_124745294 | 2.29 |
ENSMUST00000135626.8
|
Eno2
|
enolase 2, gamma neuronal |
| chr16_+_20511991 | 2.29 |
ENSMUST00000149543.9
ENSMUST00000232207.2 ENSMUST00000118919.9 |
Fam131a
|
family with sequence similarity 131, member A |
| chr9_+_102988940 | 2.28 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr4_+_104224774 | 2.26 |
ENSMUST00000106830.9
|
Dab1
|
disabled 1 |
| chr2_-_58457168 | 2.26 |
ENSMUST00000056376.12
|
Acvr1
|
activin A receptor, type 1 |
| chr4_-_25800083 | 2.25 |
ENSMUST00000084770.5
|
Fut9
|
fucosyltransferase 9 |
| chr7_+_44033520 | 2.23 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr12_+_24622274 | 2.23 |
ENSMUST00000085553.13
|
Grhl1
|
grainyhead like transcription factor 1 |
| chr7_+_18915086 | 2.22 |
ENSMUST00000120595.8
ENSMUST00000048502.10 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr7_+_36397426 | 2.21 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr7_+_130247912 | 2.21 |
ENSMUST00000207549.2
ENSMUST00000209108.2 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_+_43491389 | 2.21 |
ENSMUST00000205919.2
|
Klk5
|
kallikrein related-peptidase 5 |
| chr18_-_20247666 | 2.20 |
ENSMUST00000224432.2
|
Dsc1
|
desmocollin 1 |
| chr7_+_28988724 | 2.15 |
ENSMUST00000207714.2
ENSMUST00000048187.6 |
Ppp1r14a
|
protein phosphatase 1, regulatory inhibitor subunit 14A |
| chrX_-_23151771 | 2.14 |
ENSMUST00000115319.9
|
Klhl13
|
kelch-like 13 |
| chr4_-_43523595 | 2.14 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr19_-_7194912 | 2.12 |
ENSMUST00000039758.6
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr19_-_59931432 | 2.12 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr14_+_9646630 | 2.11 |
ENSMUST00000112658.8
ENSMUST00000112657.9 ENSMUST00000177814.2 ENSMUST00000067491.14 |
Cadps
|
Ca2+-dependent secretion activator |
| chr9_-_70048766 | 2.10 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr12_-_11200306 | 2.09 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr12_+_37930305 | 2.06 |
ENSMUST00000220990.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr18_+_37869950 | 2.05 |
ENSMUST00000091935.7
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr10_+_3316505 | 2.05 |
ENSMUST00000217573.2
|
Ppp1r14c
|
protein phosphatase 1, regulatory inhibitor subunit 14C |
| chrX_+_140258381 | 2.04 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr14_+_47121487 | 2.03 |
ENSMUST00000137543.9
|
Samd4
|
sterile alpha motif domain containing 4 |
| chr12_+_55883101 | 2.02 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr7_+_24048613 | 2.02 |
ENSMUST00000032683.6
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr9_+_37450551 | 2.01 |
ENSMUST00000002008.7
ENSMUST00000215957.2 ENSMUST00000215271.2 |
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr6_+_38639945 | 2.01 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
| chr1_+_107327374 | 2.01 |
ENSMUST00000154538.8
|
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr6_-_6882068 | 2.00 |
ENSMUST00000142635.2
ENSMUST00000052609.9 |
Dlx5
|
distal-less homeobox 5 |
| chrX_+_72546680 | 2.00 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr4_+_136013372 | 2.00 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr4_-_127251917 | 1.99 |
ENSMUST00000046498.3
|
Gjb5
|
gap junction protein, beta 5 |
| chrX_-_20816841 | 1.98 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr11_+_3438274 | 1.97 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr3_-_138780894 | 1.97 |
ENSMUST00000196280.5
ENSMUST00000200396.2 |
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chr13_-_12121831 | 1.96 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr8_-_112660363 | 1.96 |
ENSMUST00000034429.9
|
Tmem231
|
transmembrane protein 231 |
| chr11_-_69728560 | 1.94 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr18_-_12995913 | 1.94 |
ENSMUST00000121774.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr9_+_27702243 | 1.94 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr8_-_106198112 | 1.94 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr18_-_20247824 | 1.93 |
ENSMUST00000038710.6
|
Dsc1
|
desmocollin 1 |
| chr12_+_16703383 | 1.93 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr16_+_37597235 | 1.93 |
ENSMUST00000114763.3
|
Fstl1
|
follistatin-like 1 |
| chr4_-_58206596 | 1.92 |
ENSMUST00000042850.9
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr9_-_58220469 | 1.89 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr11_-_79394904 | 1.88 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr6_-_42301574 | 1.88 |
ENSMUST00000031891.15
ENSMUST00000143278.8 |
Fam131b
|
family with sequence similarity 131, member B |
| chr1_+_66507523 | 1.87 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr10_+_127337541 | 1.87 |
ENSMUST00000160019.8
ENSMUST00000160610.2 ENSMUST00000035839.3 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr2_-_164621641 | 1.86 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr7_+_18915136 | 1.86 |
ENSMUST00000144054.8
ENSMUST00000141718.8 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr8_-_107064615 | 1.86 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr3_-_104960437 | 1.85 |
ENSMUST00000077548.12
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr5_+_24305577 | 1.85 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr1_-_168259839 | 1.84 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr4_-_43523745 | 1.84 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr8_-_9821021 | 1.84 |
ENSMUST00000208933.2
ENSMUST00000110969.5 |
Fam155a
|
family with sequence similarity 155, member A |
| chr2_-_6726417 | 1.84 |
ENSMUST00000142941.8
ENSMUST00000150624.9 ENSMUST00000100429.11 ENSMUST00000182879.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr8_-_85500998 | 1.83 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr3_-_104960264 | 1.82 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr18_-_43032514 | 1.82 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_125514997 | 1.82 |
ENSMUST00000164756.4
|
Eid1
|
EP300 interacting inhibitor of differentiation 1 |
| chr12_+_75355082 | 1.81 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr17_+_47451868 | 1.81 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr13_-_78347876 | 1.81 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr15_-_77191079 | 1.80 |
ENSMUST00000171751.10
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr14_+_121272606 | 1.80 |
ENSMUST00000135010.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr9_+_107217786 | 1.80 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr15_-_13173736 | 1.79 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr4_-_140573081 | 1.78 |
ENSMUST00000026378.4
|
Padi1
|
peptidyl arginine deiminase, type I |
| chr9_+_60620272 | 1.78 |
ENSMUST00000038407.6
|
Larp6
|
La ribonucleoprotein domain family, member 6 |
| chr7_-_34914675 | 1.76 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr11_+_84848601 | 1.76 |
ENSMUST00000103194.10
|
Car4
|
carbonic anhydrase 4 |
| chr7_+_141996067 | 1.76 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr6_-_41613322 | 1.75 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr13_+_91609264 | 1.75 |
ENSMUST00000231481.2
|
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr12_-_32111214 | 1.73 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr1_-_134883645 | 1.72 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr15_-_103242697 | 1.72 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr17_-_74017410 | 1.71 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr8_-_85526972 | 1.70 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr6_-_83504756 | 1.70 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr18_-_12995395 | 1.70 |
ENSMUST00000121888.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr4_-_25800238 | 1.70 |
ENSMUST00000108199.2
|
Fut9
|
fucosyltransferase 9 |
| chrX_-_97934387 | 1.70 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr18_+_37880027 | 1.69 |
ENSMUST00000193404.2
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr6_-_83504471 | 1.67 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr2_-_172782089 | 1.67 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr3_-_138780831 | 1.67 |
ENSMUST00000029796.11
|
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chr14_-_70680659 | 1.65 |
ENSMUST00000180358.3
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr9_-_56542908 | 1.65 |
ENSMUST00000114256.2
|
Lingo1
|
leucine rich repeat and Ig domain containing 1 |
| chr5_-_24597009 | 1.65 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr3_-_33137209 | 1.65 |
ENSMUST00000194016.6
ENSMUST00000193681.6 ENSMUST00000192093.6 ENSMUST00000193289.6 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr11_+_101221895 | 1.64 |
ENSMUST00000017316.7
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr14_+_70768289 | 1.64 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr15_-_98707367 | 1.64 |
ENSMUST00000230409.2
|
Ddn
|
dendrin |
| chr14_-_70405288 | 1.64 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr9_-_30833748 | 1.62 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr3_+_75981577 | 1.61 |
ENSMUST00000038364.15
|
Fstl5
|
follistatin-like 5 |
| chr1_-_83385911 | 1.60 |
ENSMUST00000160953.8
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr19_+_10019023 | 1.59 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr5_-_138270995 | 1.59 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr10_-_120735000 | 1.58 |
ENSMUST00000092143.12
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chrX_-_20787150 | 1.57 |
ENSMUST00000081893.7
ENSMUST00000115345.8 |
Syn1
|
synapsin I |
| chr17_+_43879496 | 1.57 |
ENSMUST00000169694.2
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr7_+_45434755 | 1.57 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.9 | 5.7 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.5 | 9.0 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 1.4 | 4.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.3 | 4.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.2 | 4.7 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.2 | 3.5 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 1.1 | 3.4 | GO:0060197 | cloacal septation(GO:0060197) |
| 1.1 | 11.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.1 | 3.2 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 1.0 | 5.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 1.0 | 2.9 | GO:2000978 | positive regulation of actin filament-based movement(GO:1903116) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.0 | 2.9 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.8 | 4.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.8 | 2.5 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.8 | 1.7 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.8 | 5.8 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.8 | 5.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.8 | 2.3 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.8 | 2.3 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.7 | 2.2 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.7 | 2.8 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.7 | 2.8 | GO:1903921 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.7 | 2.7 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.7 | 2.0 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.7 | 2.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.6 | 15.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.6 | 0.6 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.6 | 2.9 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.6 | 4.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.6 | 1.2 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.6 | 2.3 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.6 | 2.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.6 | 2.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.6 | 2.2 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.5 | 2.7 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.5 | 2.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.5 | 2.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.5 | 1.9 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.5 | 4.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.5 | 5.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 0.9 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.4 | 1.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 2.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 6.6 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 1.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 2.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 1.3 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.4 | 1.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.4 | 1.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 1.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 1.9 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.4 | 1.1 | GO:0060936 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.4 | 2.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 2.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 1.4 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.3 | 1.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 1.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 1.0 | GO:0021750 | vestibular nucleus development(GO:0021750) |
| 0.3 | 2.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 5.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 2.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.3 | 2.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.3 | 1.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.3 | 1.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.3 | 1.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 4.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 1.5 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 4.5 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.3 | 1.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.3 | 0.9 | GO:0072276 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 0.9 | GO:1904414 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.3 | 2.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 1.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.3 | 2.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 | 0.8 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.3 | 1.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.3 | 4.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 1.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.2 | 1.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 2.9 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 0.7 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 | 12.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.2 | 3.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.4 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 0.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 0.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.6 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 5.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 2.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 3.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 2.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 3.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.0 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.2 | 3.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.2 | 1.5 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 0.9 | GO:1990770 | regulation of small intestine smooth muscle contraction(GO:1904347) small intestine smooth muscle contraction(GO:1990770) |
| 0.2 | 0.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 4.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.5 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.2 | 2.6 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 0.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.9 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.2 | 0.5 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 1.0 | GO:0070560 | calcium-mediated signaling using extracellular calcium source(GO:0035585) protein secretion by platelet(GO:0070560) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 | 6.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 2.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.5 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 0.5 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 2.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 1.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.2 | 0.5 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 0.2 | 3.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 0.6 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 0.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.2 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 3.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 0.5 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 1.5 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 1.4 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 1.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.7 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.1 | 2.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 5.5 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.9 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.6 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 4.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.4 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.7 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.1 | 0.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 2.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 2.6 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.2 | GO:0098910 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.1 | 1.5 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.8 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.4 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 1.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 1.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 2.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 1.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 10.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.6 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.3 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.4 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.3 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.1 | 0.3 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 1.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.7 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 4.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 6.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) regulation of rRNA processing(GO:2000232) |
| 0.1 | 2.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.6 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.3 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 2.3 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.1 | 3.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 2.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.1 | GO:0043585 | nose morphogenesis(GO:0043585) |
| 0.1 | 4.6 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 1.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 2.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 2.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.1 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 7.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 2.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.1 | 0.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 5.6 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 6.8 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 1.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.9 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 1.2 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 2.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.6 | GO:1901889 | negative regulation of cell junction assembly(GO:1901889) |
| 0.0 | 3.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0030818 | negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.5 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.7 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.0 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 2.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 3.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.8 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 2.1 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.7 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.1 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 1.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 1.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.4 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 2.0 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 2.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.9 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.0 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 2.8 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 5.3 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.9 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.1 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 1.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.2 | GO:0050942 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) positive regulation of developmental pigmentation(GO:0048087) regulation of pigment cell differentiation(GO:0050932) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.3 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0021924 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.0 | 3.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.9 | 2.6 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.8 | 3.2 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.8 | 13.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.8 | 2.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.7 | 2.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.7 | 3.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.6 | 2.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 4.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 2.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 3.7 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.5 | 7.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 1.9 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.5 | 1.4 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.4 | 4.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 2.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 2.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 5.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 2.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 0.6 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.3 | 0.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 2.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 2.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 0.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 3.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 5.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 2.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 2.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 4.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 3.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 2.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 5.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 1.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 4.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 3.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 4.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 3.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 2.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 2.4 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 22.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 7.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 3.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 3.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 7.8 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 21.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 4.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 3.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 9.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 5.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 4.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.3 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 13.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.6 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.7 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 3.1 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 15.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.4 | 4.2 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 1.4 | 4.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.1 | 9.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.1 | 3.3 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.8 | 4.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.7 | 10.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 2.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.7 | 12.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 3.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 1.8 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.6 | 2.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.6 | 4.0 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 5.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 1.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.5 | 2.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.5 | 2.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 5.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 2.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 2.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 2.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 4.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 1.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.4 | 2.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 2.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.4 | 3.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 2.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 4.5 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.4 | 1.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.4 | 3.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 1.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 5.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 3.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 3.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 1.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.3 | 1.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 3.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 2.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 2.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.7 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 1.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.6 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 1.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 2.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 5.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.6 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.2 | 0.6 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 3.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 0.6 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.2 | 2.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 2.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 3.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 2.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 0.5 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.2 | 4.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 2.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 4.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 2.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 5.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 9.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.9 | GO:0044547 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.1 | 2.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.6 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.9 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 1.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 6.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 1.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 12.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 2.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 1.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.4 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 7.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.0 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 4.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 8.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 5.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.7 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 2.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 5.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 2.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 4.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 24.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 1.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 2.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 6.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 16.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 5.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 8.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 3.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 18.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 12.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 1.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 2.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 13.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 4.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 11.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 6.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 4.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 2.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 3.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 16.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 3.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 4.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 7.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 4.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.6 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 4.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.5 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 3.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |