Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zfp410
Z-value: 0.59
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSMUSG00000042472.12 | Zfp410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp410 | mm39_v1_chr12_+_84363603_84363650 | 0.20 | 9.4e-02 | Click! |
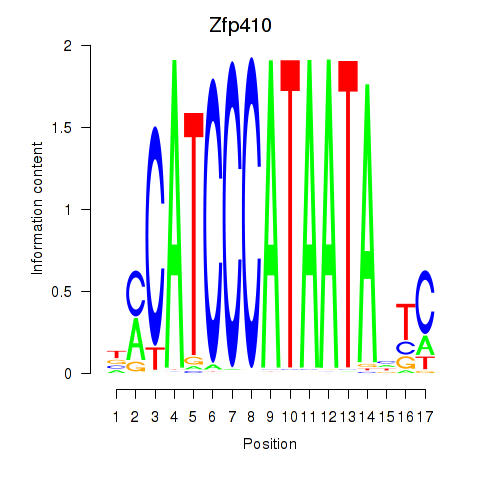
Activity profile of Zfp410 motif
Sorted Z-values of Zfp410 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp410
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_125072944 | 10.45 |
ENSMUST00000112392.8
ENSMUST00000056889.15 |
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr6_+_125073108 | 6.93 |
ENSMUST00000112390.8
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr15_-_89310060 | 4.12 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr14_-_61495934 | 3.53 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr6_+_68013772 | 3.37 |
ENSMUST00000197515.2
ENSMUST00000103315.3 |
Igkv17-121
|
immunoglobulin kappa variable 17-121 |
| chr14_-_61495832 | 3.12 |
ENSMUST00000121148.8
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr6_+_67838100 | 3.10 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr15_-_89309998 | 2.49 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr5_+_123280250 | 1.44 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr14_-_52434863 | 1.10 |
ENSMUST00000046709.9
|
Supt16
|
SPT16, facilitates chromatin remodeling subunit |
| chr5_+_31771281 | 1.02 |
ENSMUST00000031024.14
ENSMUST00000202033.2 |
Mrpl33
Gm43809
|
mitochondrial ribosomal protein L33 predicted gene 43809 |
| chr5_-_100648487 | 1.00 |
ENSMUST00000239512.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr11_-_118931731 | 0.89 |
ENSMUST00000026663.8
|
Cbx8
|
chromobox 8 |
| chr4_-_148021217 | 0.73 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr6_-_142418801 | 0.65 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr18_-_66135683 | 0.62 |
ENSMUST00000120461.9
ENSMUST00000048260.15 ENSMUST00000236866.2 |
Lman1
|
lectin, mannose-binding, 1 |
| chr18_-_44329234 | 0.60 |
ENSMUST00000239421.2
ENSMUST00000097587.5 |
Spink11
|
serine peptidase inhibitor, Kazal type 11 |
| chr5_-_104125192 | 0.53 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125270 | 0.53 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chrX_-_72442342 | 0.51 |
ENSMUST00000180787.3
|
Gm18336
|
predicted gene, 18336 |
| chr5_-_104125226 | 0.50 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr14_-_56299764 | 0.47 |
ENSMUST00000043249.10
|
Mcpt4
|
mast cell protease 4 |
| chr4_-_148021159 | 0.39 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr16_-_21606546 | 0.38 |
ENSMUST00000023559.7
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| chr6_+_40419797 | 0.31 |
ENSMUST00000038907.9
ENSMUST00000141490.2 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chr7_+_43865369 | 0.26 |
ENSMUST00000074359.4
|
Klk1b5
|
kallikrein 1-related peptidase b5 |
| chr7_+_12712139 | 0.20 |
ENSMUST00000108536.3
|
Zfp446
|
zinc finger protein 446 |
| chr5_-_151510389 | 0.02 |
ENSMUST00000165928.4
|
Vmn2r18
|
vomeronasal 2, receptor 18 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 17.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 1.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 6.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.6 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 6.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 6.7 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.6 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.6 | 17.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 6.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 17.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 0.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 17.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 6.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |